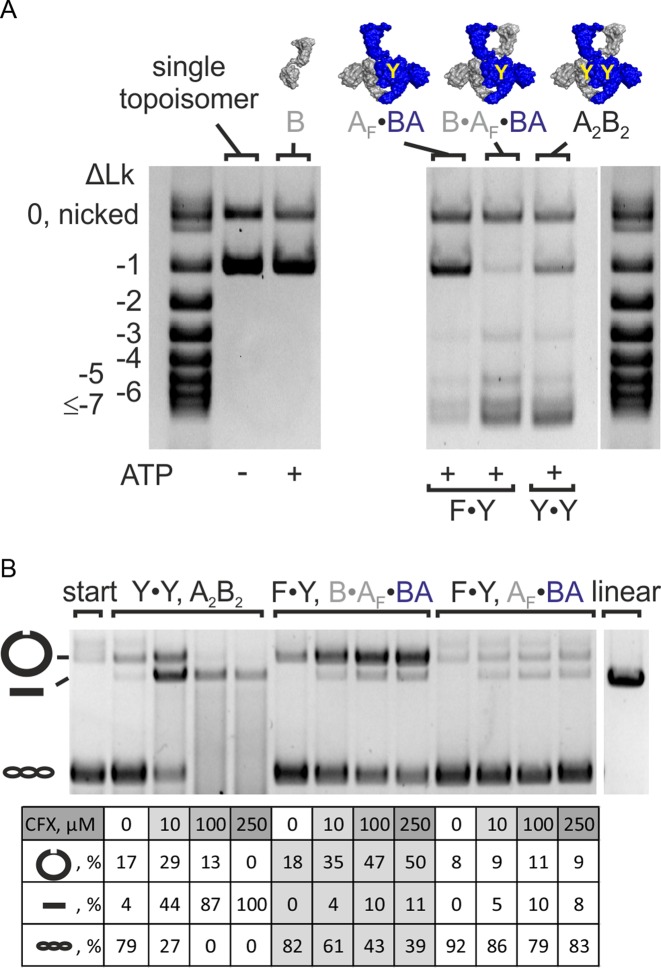
Figure 3.
Change in linking number and DNA cleavage. (A) Supercoiling of a single topoisomer (ΔLk = −1, right) by gyrase containing one (F·Y, B·AF·BA) or two (Y·Y, A2B2) catalytic tyrosine(s). Both enzymes change the linking number in steps of two. DNA supercoiling was performed for 4 min at 37°C. Concentrations were 20 nM GyrB, 20 nM GyrA and 20 nM GyrB (corresponding to 10 nM A2B2) or 25 nM AF·BA and 100 nM GyrB (corresponding to 25 nM BA·AF·B), respectively. Gyrase with two (A2B2) and one catalytic tyrosine(s) (B·AF·BA) introduces negative supercoils in steps of two. The AF·BA heterodimer serves as a negative control. AF·BA does not supercoil DNA, confirming that the observed supercoiling activity of B·AF·BA is not caused by contaminations in the preparation. (B) DNA cleavage in the presence of CFX. B. subtilis gyrase (A2B2) causes nicking and linearization, corresponding to single- and double-strandP cleavage. Gyrase containing only one catalytic tyrosine (B·AF·BA) shows an accumulation of nicked DNA, in agreement with single-strand cleavage. CFX concentrations are 0–250 μM (arrow). The gyrase:DNA ratio is 5:1. The table below the gel summarizes the fractions of nicked, linear and supercoiled DNA according to densitometric analysis. linear: pUC18 linearized by BamHI; start: DNA substrate; A2B2: heterotetrameric gyrase; A2: GyrA dimer; AF: cleavage-deficient GyrA subunit (GyrAY123F); B: GyrB; BA: GyrBA fusion protein; BAΔCTD: GyrBA fusion protein lacking the GyrA CTD; ΔLk: linking number difference.