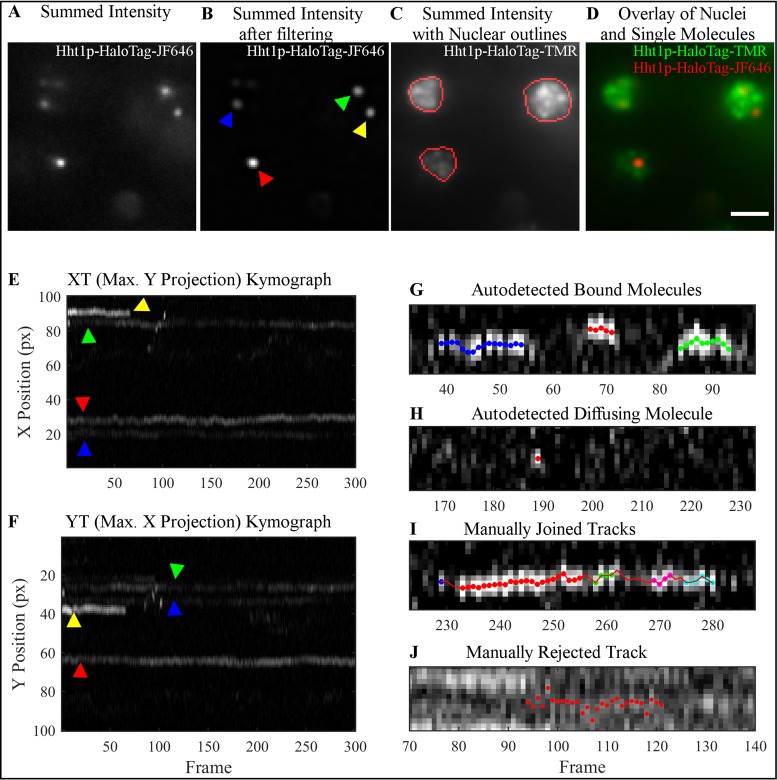
Figure 2.
Overview of the steps involved in kymograph-based tracking of single molecules of Hht1p-HaloTag labeled with JF646. Imaging is done on HILO microscope. (A) Projection of the intensity summed over all 300 frames of a representative movie to visualize all particles in the 647 nm channel. (B) Same projection as in (A) after noise reduction with the band-pass filtering module of the software. (C) Summed intensity projection of the same field-of-view as in (A) in the 561 nm channel to visualize nuclei. Red outlines indicate user-identified ROIs in which particle tracking is performed. (D) Overlay of 647 nm (red) and 561 nm (green) channels demonstrating particles residing in the nuclei. (E and F) Kymographs XT, (E) Y-projections, and YT, (F) X-projection of all 300 frames from the movie shown in (B) after filtering. Both kymographs are used for manual correction of tracks because apparent track overlap in one dimension resulting from particles in different nuclei are typically well-separated in the other dimension. Colored arrows indicate the corresponding particles shown in the summed image displayed in (B). Examples of (G and H) correctly autodetected and (I and J) manually edited tracks shown as kymographs. (G) The software successfully identifies tracks that are continuous or contain gaps of a single frame. Red, green and blue spots indicate three separate tracks from this image. (H) The algorithm also identifies freely diffusing molecules, which are characterized by a bright spot in a single frame with dimmer (usually not autodetected) spots in the preceding and/or following frames. (I) If a track contains gaps of several frames, the user must manually join the autodetected track segments. (J) Over-crowding makes it challenging to follow a single particle, and so these types of tracks are typically thrown out. See text for our recommendations on when to merge track segments as in (I), or leave them separated as in (G).