Figure 2.
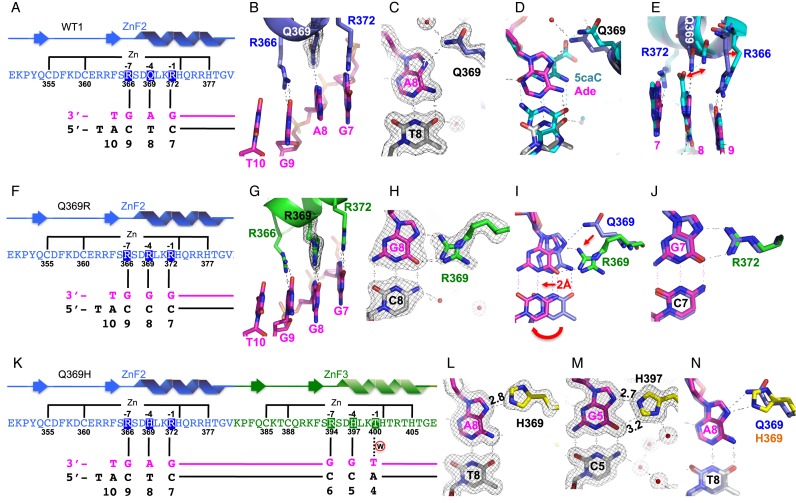
Structural analysis of purine (adenine or guanine) binding in the 3′ triplet. (A–E) Structure of normal WT1 in complex with the A:T base pair. (A) Schematic representation of the ZF2 (from N-to-C termini) and the recognition sequence (from 3′-to-5′ of the top strand). The complementary strand (bottom) has a 5′ overhanging thymine used for the crystallization study. (B) R372, Q369, and R366 of ZF2 (in blue) interact, respectively, with Gua7, Ade8, and Gua9 of DNA (in magenta). (C) Q369-Ade8 interaction. The 2Fo-Fc electron density, contoured at 1σ above the mean, is shown in gray. (D) Structural superimposition of Q369-Ade (in blue for Q369, magenta for Ade, and gray for Thy) and Q369-5caC (in cyan; PDB: 4R2R). (E) The side chains of Q369 and R366 displaced shifts from binding of 5caC (in cyan) to Ade (in magenta). (F–J) Structure of Q369R in complex with the G:C base pair. (F) Schematic representation of the ZF2 with Q369R mutant. (G) The three arginines (R372, R369, and R366) align with three adjacent guanines along the DNA major groove. (H) R369-Gua8 interaction. (I) Structural superimposition of Q369-Ade (in blue) and R369-Gua (in green for R369 and magenta for Gua). (J) R372-Gua7 interaction. (K–N) Structure of Q369H in complex with the A:T base pair. (K) Schematic representation of the ZF2 (Q369H mutant) and ZF3 in recognition of the corresponding triplets. (L) H369-Ade8 interaction. (M) H397-Gua5 interaction. (N) Structural superimposition of Q369-Ade (in cyan) and H369-Ade (yellow for H369, magenta for Ade8).