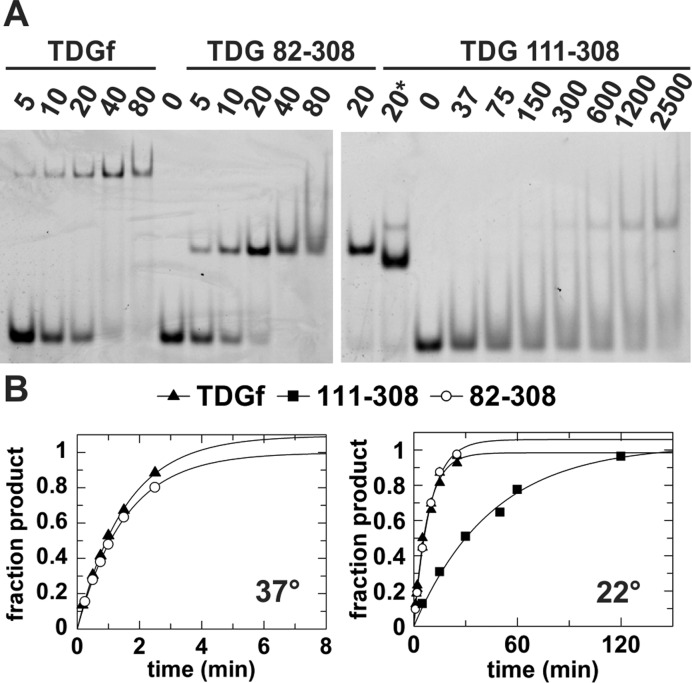
Figure 2.
Biochemical studies of full length TDG, TDG82-308 and TDG111-308. (A) Equilibrium binding of a given TDG construct to DNA (10 nM) containing a G·TF mismatch, where TF is a non-cleavable Thd analog, monitored by electrophoretic mobility shift assays (EMSA). The concentration of enzyme (nM) is indicated. For TDG111-308, the left lane marked ‘20*’ indicates binding to abasic DNA product (10 nM, 28 bp), to show the mobility of a tight 1:1 complex, given that such a complex is not clearly observed for G·TF DNA in the gel. (B) Single-turnover kinetics for excision of T from a G·T DNA substrate by the three TDG constructs, at 37°C and 22°C (TDG111-308 not stable at 37°C). Rate constants are kmax = 0.651 ± 0.044 min−1 for TDG and kmax = 0.655 ± 0.020 min−1 for TDG82-308 at 37°C, and kmax = 0.126 ± 0.008 for TDG, kmax = 0.108 ± 0.006 min−1 for TDG82-308, and kmax = 0.022 ± 0.002 min−1 for TDG111-308 at 22°C.