Abstract
Sodium is one of the most abundant metals in the environment and in biology, playing critical ecological and physiological roles. Na+ is also the most common buffer salt for nucleic acids research, while its specific interaction with DNA has yet to be fully studied. Herein, we probe a highly selective and robust Na+ aptamer using 2-aminopurine (2AP), a fluorescent adenine analog. This aptamer has two DNA strands derived from the Ce13d DNAzyme. By introducing a 2AP at the cleavage site of the substrate strand, Na+ induces ∼40% fluorescence increase. The signaling is improved by a series of rational mutations, reaching >600% with the C10A20 double mutant. This fluorescence enhancement suggests relaxed base stacking near the 2AP label upon Na+ binding. By replacing a non-conserved adenine in the enzyme strand by 2AP, Na+-dependent fluorescence quenching is observed, suggesting that the enzyme loop folds into a more compact structure upon Na+ binding. The fluorescence changes allow for Na+ detection. With an optimized sequence, a detection limit of 0.4 mM Na+ is achieved, reaching saturated signal in less than 10 s. The sensor response is insensitive to ionic strength, which is critical for Na+ detection.
INTRODUCTION
Over the past two decades, many DNA sequences have been discovered to selectively recognize an impressive number of metal ions (1–6). The majority of such sequences are DNAzymes that require metals for activity. Most previous work focused on divalent metals such as Pb2+ (7,8), Zn2+ (9,10), Cu2+ (11), UO22+ (12,13), Cd2+ (14) and Hg2+ (15,16). Recently, DNAzymes sensitive to trivalent lanthanides have also been reported (17–20). Aside from Ag+ (21,22), binding of monovalent cations such as K+ (23) and Tl+ (24) has mainly relied on G-quadruplex DNA.
Sodium is one of the most abundant and important metals in the environment and in biology. Its specific interaction with DNA, however, has not been fully explored. Na+ is always treated as a diffusive charge around DNA without considering its specific chemical properties (25). Recently, a few Na+-specific RNA-cleaving DNAzymes were discovered (19,26–28). For example, Lu et al. reported the NaA43 DNAzyme reaching a rate of 0.1 min−1 with 400 mM Na+ alone (26). Interestingly, the NaA43 has a high sequence analogy to a lanthanide-dependent DNAzyme named Ce13d we reported (19,29). Further assays indicate that Ce13d also requires Na+ (30,31), suggesting the possibility of a common Na+ aptamer in these DNAzymes.
The Na+ binding property of the Ce13d has been probed by activity assay, dimethyl sulfate (DMS) footprinting and sensitized Tb3+ luminescence (29–32). However, little is known about the folding of the Na+ aptamer in this DNAzyme. Metal-induced folding is an important aspect of understanding aptamers and DNAzymes (33–39). In this work, we aim to address the following questions: does Na+ induce a specific folding of the DNAzyme, and can we design a folding based Na+ sensor?
Since Na+ is a cation and DNA is a polyanion, Na+ also promotes non-specific DNA condensation. Therefore, it is critical to distinguish specific DNA folding from non-specific interactions. Instead of using techniques that probe global folding of DNA, such as fluorescence resonance energy transfer, studying local folding is more appropriate here. 2-aminopurine (2AP) is a fluorescent adenine analog (40). Its emission intensity is strongly affected by the local base stacking environment (40). Such a property has made 2AP a powerful probe for nucleic acids (40–42), and related analytical applications (43–45). In this work, we probe the Ce13d using 2AP at a few different positions, allowing us to map its full folding picture.
MATERIALS AND METHODS
Chemicals
The ribo-2AP-modified substrate was from Trilink BioTechnologies (San Diego, CA, USA). The rest of the DNA samples were purchased from Integrated DNA Technologies (Coralville, IA, USA), and their sequences and modifications are shown in Supplementary Table S1. The metal salts were from Sigma-Aldrich, and buffers were from Mandel Scientific Inc. (Guelph, Ontario, Canada). Milli-Q water was used to prepare all the buffers and solutions.
Fluorescence spectroscopy
The DNAzyme complexes were annealed in buffer A (100 mM LiCl, 50 mM Tris, pH 7.5) by heating the samples to 80°C for 2 min and then gradually cooling to 4°C over 30 min. When 2AP was modified at the substrate strand, the complex was prepared with 1 μM substrate and 2 μM enzyme. When the 2AP was at the enzyme strand, 2 μM substrate and 1 μM enzyme were used. The 2AP emission was measured immediately after adding metal using a fluorometer (FluoroMax-4, Horiba Scientific) by exciting at 310 nm. The spectra were recorded from 360 to 450 nm, and the peak intensity at 370 nm was used for quantification. The Kd value was calculated based on: 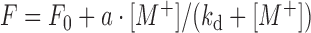 , where F0 and F are the fluorescence intensity before and after adding metal; [M+] is the metal concentration; and a is the fluorescence change when [M+] = ∞.
, where F0 and F are the fluorescence intensity before and after adding metal; [M+] is the metal concentration; and a is the fluorescence change when [M+] = ∞.
Cleavage activity assays
The DNAzyme complex was annealed in buffer C (25 mM NaCl, 50 mM MES, pH 6.0) with 1 μM FAM-labeled substrate strand and 2 μM enzyme. At room temperature, 10 μM Ce3+ was added to initiate the cleavage reaction. At each designated time point, a 2.5 μl aliquot was mixed with 14 μl urea (8 M) to quench the reaction. The cleavage products were separated using 15% denaturing polyacrylamide gel electrophoresis (dPAGE) and analyzed by a ChemDoc MP imaging system (Bio-Rad).
RESULTS AND DISCUSSION
Na+-induced folding of the Ce13d
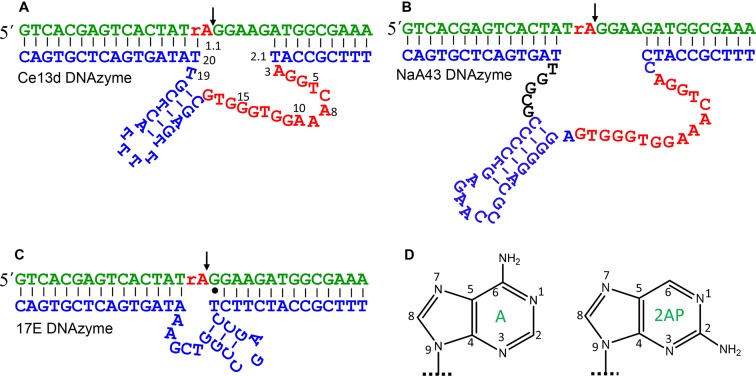
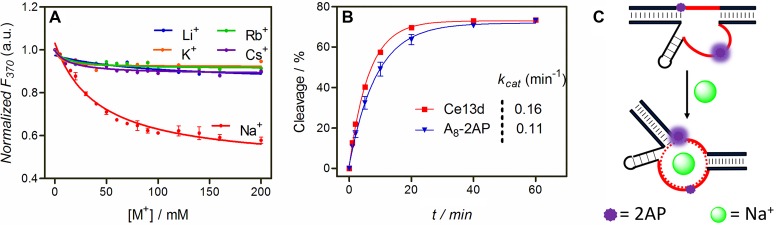
The secondary structures of the Ce13d and NaA43 DNAzymes are shown in Figure 1A and B, respectively. They have the same substrate (in green) containing a single ribo-adenine (rA) as the cleavage site. The NaA43 is active with Na+ alone, while the Ce13d requires both Na+ and a lanthanide ion such as Ce3+. They have 16 identical nucleotides related to Na+ binding (in red) (29,30). A discussion regarding their secondary structures is shown in Supplementary Figure S1. Our goal here is to probe Na+-induced DNA folding and exploit a folding-based Na+ sensor. Out of these two DNAzymes, we focused on the Ce13d for the following reasons. First, Ce13d is inactive with Na+ alone, allowing us to study Na+-dependent folding independent of cleavage. Without cleavage, the Ce13d acts as a Na+ aptamer. Second, the Ce13d is smaller in size. Third, Ce13d appears to bind Na+ more strongly (vide infra). Since 2AP is an adenine analog (Figure 1D), we first replaced the cleavage site rA1.1 by a 2AP. We hypothesize that Na+ binding might affect base stacking at this position, which can be reflected by the 2AP fluorescence.
Figure 1.
The secondary structures of (A) the Ce13d, (B) the NaA43 and (C) the 17E DNAzymes. The sequences highlighted in red are identical in the Ce13d and NaA43. The cleavage sites are marked by the arrowheads. (D) The structures of adenine and 2AP.
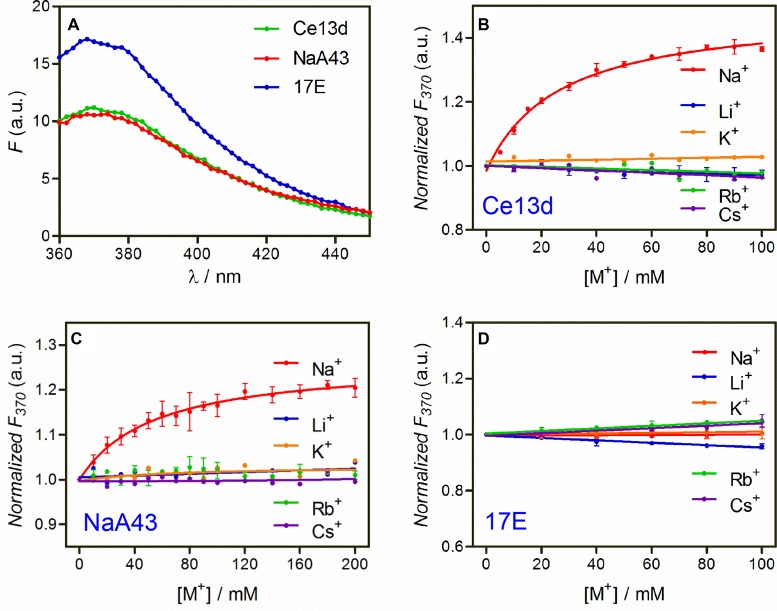
With a 2AP ribonucleotide at the rA1.1 position, the Ce13d complex emits at 370 nm (Figure 2A, green trace). We then titrated the chloride salts of various monovalent metals. With increasing Na+ concentration, the fluorescence gradually increased with a dissociation constant (Kd) of 27 mM Na+ (Figure 2B, red trace). This value is quite similar to those obtained using sensitized Tb3+ luminescence and DMS footprinting (30). This fluorescence increase can be interpreted as a loss of 2AP base stack due to folding of the aptamer by Na+. In contrast, K+ failed to induce a fluorescence change, while the rest of the group 1A metals resulted in a slight signal drop. This indicates that Na+ can selectively fold the Ce13d, and the 2AP modification does not alter its Na+ specificity. The specific Na+ binding of the 2AP-modified DNAzyme is also supported by a sensitized Tb3+ luminescence experiment (Supplementary Figure S2). While Na+ binding was confirmed, the signal enhancement was quite poor. With 100 mM Na+, the emission increased by only ∼40%. It needs to be emphasized that Ce3+ was not added in our 2AP fluorescence experiments. The roles are Ce3+ and Na+ are relatively independent in the Ce13d: Ce3+ binds to the scissile phosphate to assist cleavage (29), while Na+ binds to the nucleotides in red in Figure 1A. Without Ce3+, no cleavage takes place (Supplementary Figure S3), and only DNA folding is probed (30).
Figure 2.
(A) The initial fluorescence spectra of the Ce13d, NaA43 and 17E DNAzyme complexes with a 2AP replacing the rA1.1 position in the absence of Na+. Relative fluorescence intensity at 370 nm of the 2AP-modified (B) Ce13d, (C) NaA43 and (D) 17E DNAzyme as a function of salt concentration. All the complexes were formed in buffer A (100 mM LiCl, 50 mM Tris, pH 7.5) with 1 μM 2AP-modified substrate strand and 2 μM enzyme. The dilution effect during titration was corrected. The error bars in all the figures represent the standard deviation from at least two independent measurements.
Having confirmed that 2AP is an appropriate probe for the Ce13d, we next tested the NaA43 DNAzyme using the same 2AP-labeled substrate (see Figure 1B for structure). Na+-specific fluorescence enhancement was also observed (Figure 2C), suggesting a similar Na+ binding. However, the signal increase was even lower (only ∼20%) with a Kd of 54 mM Na+, which is consistent with the apparent Kd from its cleavage activity (40 mM) (26). Since the NaA43 is active with Na+ alone, to avoid potential cleavage by the NaA43, we also used the non-cleavable full-DNA 2AP substrate (named Sub-deoxyribo-2AP), and the amount of fluorescence increase remained similar (Supplementary Figure S4).
Since the Ce13d and NaA43 are identical in the nucleotides marked in red, the lower Na+ affinity of NaA43 is likely due to the few nucleotides on the other side of the hairpin (Figure 1B, in black). These nucleotides play a critical catalytic role (e.g. replacing the role of Ce3+ in Ce13d) (30). As discussed below, these nucleotides may modulate the amount of 2AP signal change by affecting the initial or final base stacking environment of the 2AP. The larger size of NaA43 may also make the aptamer more floppy and affect its Na+ binding.
We then did a control experiment with the 17E DNAzyme (see Figure 1C for structure), which is a well-studied system with no Na+ specificity (46). In this case, the fluorescence failed to increase with any monovalent metal (Figure 2D), further supporting the unique property of the Ce13d and NaA43 for specific Na+ binding. It is interesting to note that the initial fluorescence is stronger in 17E (Figure 2A), suggesting its 2AP is initially less stacked. Although these three DNAzymes all have a similar overall secondary structure, as illustrated below, the 2AP emission is more sensitive to its local environment instead of the global folding of the whole DNA.
Tuning the local base pairing environment
While this 2AP-modified Ce13d behaves like a well-defined Na+ aptamer, the saturated signal increase was only ∼40%. To improve sensitivity and gain further insights, we aimed to tune the local stacking of the 2AP. Like adenine, 2AP can also base pair with thymine (Figure 3A). We chose to focus on the T20 position (Figure 3A, lower panel), since it may directly interact with the 2AP label via Watson–Crick base pairing. Note in our previous papers (19,29), this rA1.1 position (now replaced with the 2AP) was thought to be non-paired as predicted by Mfold (47).
Figure 3.
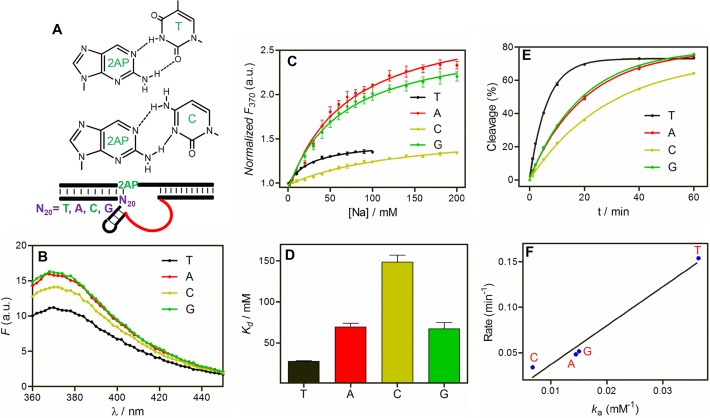
(A) The base pair structures of 2AP with thymine and cytosine (upper panels), and a cartoon showing the Ce13d mutants at the N20 position paired with the 2AP label (lower panel). (B) The initial fluorescence spectra of the N20 mutants of Ce13d DNAzyme in absence of Na+. (C) Relative fluorescence increase of the Ce13d N20 mutants as a function of Na+ concentration. (D) Kd of each mutant calculated from (C). (E) Cleavage kinetics of the mutants using the cleavable substrate (without 2AP label) in the presence of Ce3+. (F) Correlation between cleavage rate and Ka of the mutants. The 2AP fluorescence was measured in buffer A, with 1 μM 2AP modified complex. The cleavage assays were carried out in buffer C (25 mM NaCl, 50 mM MES, pH 6.0), with 1 μM FAM-labeled rA substrate, 2 μM enzyme and 10 μM Ce3+.
We respectively mutated the T20 position to A, C and G. The initial fluorescence intensity of these mutants followed the order of A ≅ G > C > T (Figure 3B), which agrees with their 2AP base pairing affinity ranking (e.g. T-2AP has the strongest affinity and thus the weakest initial fluorescence) (48,49). The schemes of 2AP pairing with T and C are shown in Figure 3A. We interpret an increased 2AP emission as relaxed base stacking (40,49). Since a stronger 2AP base pair results in a weaker initial fluorescence, this result also supports that the 2AP pairs with T20.
After understanding their initial states in the absence of Na+, we next titrated Na+. Interestingly, Na+-dependent fluorescence increase still occurred for all the mutants (Figure 3C). In particular, the highest enhancement was observed with A and G at the N20 position. Note that A and G had the highest initial fluorescence, yet they still yielded the highest relative enhancement (∼130% compared to 34% for T and C). This might be explained by T20 and C20 still partially base paired with the 2AP even after Na+ binding, thus limiting their signal increase.
The data in Figure 3C were fitted to Na+ binding curves to extract the Kd values (Figure 3D). Interestingly, the original Ce13d with a T at the N20 position has the lowest Kd (i.e. the tightest binding), while a C has the highest Kd, leaving A and G in between. Therefore, the Na+ binding affinity and the amount of fluorescence enhancement do not correlate.
We also measured the cleavage activity of each mutant using the rA substrate plus Ce3+ (Figure 3E). The highest activity was observed with a T at the N20 position, followed by A/G and then C. A good correlation exists between the Ka values of Na+ (1/Kd) and the cleavage rates (Figure 3F). Therefore, Na+ binding affinity determines its cleavage activity. This suggests that Na+ binding might be a pre-requisite of the cleavage reaction. A higher binding affinity, however, is not necessarily correlated with a higher fluorescence change, since the latter is determined by the local base pairing of the 2AP label. This set of experiments significantly enhanced our understanding of Na+ binding, cleavage activity and folding of Ce13d. From the analytical standpoint, we have discovered mutations with higher Na+ sensitivity.
Mutating the Na+ binding loop
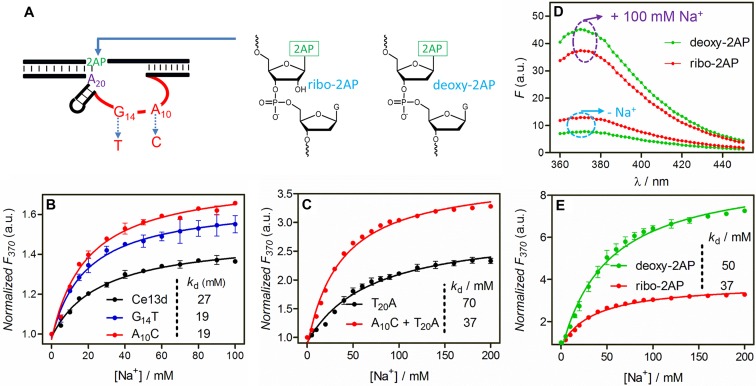
After studying the N20 position on the small loop in the enzyme strand, we turned our attention to the large loop (Figure 1A in red). This loop is highly conserved, and only a few nucleotides can be mutated without significantly undermining the cleavage activity, such as the A3 and G14 (29). Our previous Tb3+ luminescence studies also discovered a few mutants with a similar or even better Na+ binding affinity than the original Ce13d, such as the G14T and A10C mutants (e.g. A10C means the N10 position A is mutated to C; Figure 4A) (32). We also hybridized a few such active mutants with the 2AP substrate and titrated them with Na+. Among them, the A10C and G14T mutants displayed the strongest signal (Supplementary Figure S5), stronger than that of the original Ce13d. Their Kd's (∼19 mM) were lower as well (Figure 4B). Such larger fluorescence change and tighter Na+ binding are beneficial for Na+ sensing.
Figure 4.
(A) Three positions in the Ce13d DNAzyme were studied in this part: A10 and G14 in the enzyme strand and the 2AP label in the substrate. The schemes of ribo-2AP and deoxyribo-2AP are shown. (B) Relative fluorescence increase at 370 nm of the wild-type Ce13d, and its mutants as a function of Na+ concentration. (C) Normalized fluorescence at 370 nm of the T20A mutant and the A10C plus T20A double mutant as a function of Na+ concentration. (D) Fluorescence spectra of the double mutant with the ribo-2AP or deoxyribo-2AP substrate before and after adding 100 mM Na+. (E) Relative fluorescence increase at 370 nm of the double mutant with the ribo-2AP or deoxyribo-2AP substrate as a function of Na+ concentration. All the experiments were performed in buffer A.
A highly sensitive double mutant
The above studies indicate that the T20A mutant yields higher signal increase (∼130%), but it has slightly decreased Na+ binding affinity. On the other hand, mutating the Na+ binding loop (e.g. A10C) results in higher Na+ binding affinity without significantly improving the signaling sensitivity. We reason that by making double mutants, one at each position, both tighter binding and larger signal change might be achieved. To test this idea, we mutated T20A and A10C together. Indeed, this double mutant has a much higher fluorescence increase of 230% (Figure 4C) compared to the single mutants in Figure 4B. The Kd of this double mutant (37 mM Na+) is also better than the T20A single mutant. Overall, this double mutant is the best for Na+ signaling so far in terms of binding affinity and sensitivity.
Deoxyribo-2AP is even more sensitive
To minimize perturbation, we have been using a ribo-2AP in the substrate strand so far. We were also interested in testing the deoxyribo-2AP at the A1.1 position for even higher stability (see Figure 4A for their structures). Next, we hybridized the double mutant with the deoxyribo-2AP substrate. Interestingly, its background in the absence of Na+ was 41% lower compared to its RNA analog (Figure 4D, lower green trace). The only difference between these two is the 2′-OH group. This suggests that the deoxyribo-2AP has a better initial base stacking. Interestingly, after adding 100 mM Na+, the deoxyribo-2AP achieved an even stronger final fluorescence (Figure 4D, upper green trace).
Next, we performed a careful Na+ titration (Figure 4E). Compared to the ribo-2AP sample, the deoxyribo-2AP signal drastically increased with Na+ concentration, reaching more than 630% fluorescence enhancement, about 3-fold higher than that with the ribo-2AP sample. The fact that Ce13d can achieve this must suggest a quite large change in the local base pairing environment.
Probing local folding at the enzyme loop
In all the above studies, the 2AP label was placed at the A1.1 position in the substrate strand. This has allowed us to mutate the enzyme strand for mechanistic understanding and for improving signaling. Next, we want to probe the folding of the enzyme strand. The conserved loop contains four adenine resides: A3, A8, A9 and A10. Based on our previous mutation studies, A3 and A8 are unimportant for activity and can be mutated to other nucleotides while still retaining high activity (i.e. rate decreases by less than 10-fold for most mutants) (29). Some mutations even retained full activity (e.g. mutating A3 or A8 to guanine). On the other hand, any mutations to A9 or A10 nearly fully abolished the activity. Since A8 is in the middle of the Na+ binding loop, we first replaced it by 2AP to probe the aptamer folding at this position (named Ce13d-A82AP). In this case, the substrate strand was a non-labeled full-DNA analog.
Interestingly, upon titrating Na+ to the Ce13d-A82AP complex, the 2AP fluorescence gradually decreased by ∼40% (Figure 5A), while other cations did not have much effect. Therefore, this fluorescence decrease is also attributed to Na+-induced aptamer folding. Using the cleavable RNA substrate, the cleavage activity of the Ce13d-A82AP mutant is largely retained, with the rate being comparable to that of the original Ce13d (Figure 5B). This control also supports that we are probing the folding relevant to the original DNA.
Figure 5.
(A) Normalized fluorescence intensity at 370 nm of the Ce13d-A82AP as a function of monovalent salt concentration. (B) Cleavage kinetics by the original Ce13d and the Ce13d-A82AP mutant. The assay was performed in Buffer C with 10 μM Ce3+. (C) A scheme showing Na+-induce folding of the Ce13d. Na+ binding enhances the 2AP emission at the A1.1 position (suggesting relaxed base stacking) and quenches the 2AP emission at the A8 position (enhanced base stacking).
We also labeled a 2AP at the A9 position. In this case, Na+ and Li+ behaved similarly without yielding much signal change (Supplementary Figure S6). It is likely that this modification has fully disrupted Na+ binding, which is consistent with this position being highly conserved in the original DNAzyme. Since A8 and A9 are right next to each other, this result also supports that our observed 2AP fluorescence change is due to specific Na+ binding instead of non-specific interactions.
A model of Na+-induced Ce13d folding
Our data explicitly indicate a specific folding of the Ce13d upon Na+ binding. By probing local folding at two different positions, one in the substrate and the other in the enzyme, we put forward a model describing this folding in Figure 5C. The binding of Na+ relaxes the stacking between A1.1 and its neighbor bases. Therefore, this position goes from a confined state to a more flexible state upon Na+ binding. On the other hand, the A8 position becomes more stacked after binding Na+, suggesting adaptive folding of this loop typical of aptamer binding (50). Although two 2AP fluorophores are drawn in Figure 5C, we only labeled them one at a time in this work. This scheme is to put our data together to understand the overall folding.
We previously characterized Na+ binding by Ce13d using DMS footprinting (30), which probes only the guanine residues in this conserved loop. In addition, sensitized Tb3+ luminescence was performed that probes the overall global folding. The 2AP information here complements the previous studies by providing a more detailed picture of Na+-induced folding at the important adenine positions.
It needs to be noted that while many metal-specific DNAzymes have been reported (e.g. the GR5 DNAzyme for Pb2+ (7), the 17E DNAzyme for Pb2+ (51,52) the 39E for UO22+ (12) and some lanthanide-specific DNAzymes (17,18,20)), the Ce13d and NaA43 are the first examples of having a well-defined metal binding aptamer domain. For most other DNAzymes, metal specificity might not come from typical aptamer-type of metal binding. Despite extensive efforts, finding metal aptamers in most DNAzymes has been elusive (38,53,54).
Selecting aptamers for metal ions has been a long-standing challenge due to difficulties associated with metal ion immobilization. So far, few high-performance metal aptamers were isolated using selection methods. While it is possible to immobilize the DNA library (55) or using emulsion droplets to bypass metal immobilization (56), this work suggests an alternative method to obtain aptamers via DNAzyme selection. In this method, no immobilization of metal ions or the library, or the use of emulsion is needed.
Highly sensitive, selective and robust Na+ sensing
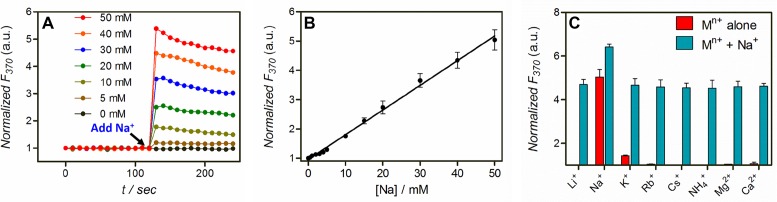
Na+ is a highly important analyte both in the environment and in biology. With very limited chemical features for rational ligand design, however, Na+ detection has been a long-standing challenge. While in this case Na+ can be measured based on DNAzyme cleavage (26), folding-based sensors are reversible allowing continuous sensing. Herein, the double mutant with the deoxyribo-2AP substrate was tested as a Na+ sensor. First, we measured the kinetics of signaling at different Na+ concentrations. The fluorescence change was completed within 10 s (the shortest time possible for manual pipetting) at all tested Na+ concentrations (Figure 6A). We further plotted the fluorescence enhancement in the range of 0–50 mM Na+, and a linear relationship was obtained (Figure 6B). From this curve, a detection limit of 0.4 mM was obtained from 3σ/slope (σ is the background variation of the blank). This figure of merit is similar to that of the cleavage-based sensor (26).
Figure 6.
(A) Kinetics of fluorescence change of the optimized Na+ sensor containing the double mutant and the deoxyribo-2AP substrate at different Na+ concentrations. (B) A linear response of the sensor is observed with up to 50 mM Na+. (C) The sensor specificity test with various monovalent (50 mM) and divalent (5 mM) metal ions alone, and with additional 50 mM Na+.
We then measured the sensor specificity against various monovalent and divalent metal ions. Among the tested ions, only K+ shows a weak response while other ions are silent, suggesting that the sensor is highly selective for Na+ (Figure 6C, red bars). In addition, the sensor shows a similar response to Na+ in the presence of 50 mM monovalent or 5 mM divalent background metal ions (Figure 6C, blue bars). Therefore, this sensor works in different ionic strength conditions, and it resists ionic strength variation. This is a highly important property for DNA-based metal sensing as discussed previously. Even in the presence of 100 mM crown ether (18-crown-6) that can bind Na+, this sensor still showed a similar performance (Supplementary Figure S7). We compared our sensor performance with previously reported Na+ sensors in Table 1, and our sensor has the best overall performance.
Table 1. A comparison between our Na+ aptamer sensor and representative crown ether based sensors.
| Sensors | LOD (mM Na+) | Selectivity over K+ | Selectivity over Li+ | Ref. |
|---|---|---|---|---|
| This work | 0.4 | 10-fold | >100-fold | |
| Crown ether | 0.1 | 15-fold | ∼4-fold | (57) |
| Crown ether | <2.7 | ∼10 | ∼2-fold | (58) |
| Crown ether | <5 | 3.4-fold | NA | (59) |
NA = No data available.
Finally, we tested this sensor in measuring Na+ in the Atlantic Ocean water. Using the standard addition method, the Na+ concentration in ocean sample was determined to be 276 mM (Supplementary Figure S8A). Within the error range, this result is identical to the value quantification measured by ICP (Supplementary Figure S8B).
CONCLUSIONS
In summary, we studied the local folding of a Na+ binding aptamer using 2AP as a fluorescence probe. The following important findings were made in this work. First, 2AP was modified at both the substrate strand by replacing the cleavage site adenine base and at the enzyme strand by replacing a replaceable adenine. In both cases, the 2AP fluorescence specifically changed with Na+ titration (but in opposite directions), while other ions had little effect on fluorescence. This work also gives a vivid picture of the aptamer conformational change upon Na+ binding, providing important mechanistic insights for future structural biology studies. In addition, the effect of important nucleotides in the enzyme strand were individually probed, providing insights into their roles in metal binding and catalysis. This work explicitly demonstrates a highly robust DNA aptamer. This is the first example of extracting a metal binding aptamer resulted from DNAzyme selections. Most previous DNAzymes can achieve high metal specificity for their activity, yet the origin of such specificity is often unknown and cannot be related to a metal-binding aptamer. Finally, we identified a double mutant with the deoxyribo-2AP label, which allows >600% fluorescence enhancement relative to the background. A highly sensitive folding-based Na+ sensor was demonstrated with a detection limit of 0.4 mM Na+.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Waterloo, the Natural Sciences and Engineering Research Council of Canada (NSERC); Foundation for Shenghua Scholar of Central South University and the National Natural Science Foundation of China [21301195]; Fellowship from the China Scholarship Council (CSC) [201406370116 to W.Z.]. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X.-B., Kong R.-M., Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Y., Lu Y. DNA as sensors and imaging agents for metal ions. Inorg. Chem. 2014;53:1925–1942. doi: 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman S.K. Catalytic DNA: Scope, applications and biochemistry of deoxyribozymes. Trends Biochem. Sci. 2016;41:595–609. doi: 10.1016/j.tibs.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y. New transition metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem. Eur. J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Schlosser K., Li Y.F. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Breaker R.R., Joyce G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 8.Hwang K., Wu P., Kim T., Lei L., Tian S., Wang Y., Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem., Int. Ed. 2014;53:13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoro S.W., Joyce G.F., Sakthivel K., Gramatikova S., Barbas C.F. III. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 10.Li J., Zheng W., Kwon A.H., Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang P.-J.J., Liu J. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal. Chem. 2016;88:3341–3347. doi: 10.1021/acs.analchem.5b04904. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Brown A.K., Meng X., Cropek D.M., Istok J.D., Watson D.B., Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu P., Hwang K., Lan T., Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P.-J.J., Liu J. Rational evolution of Cd2+-specific dnazymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015;43:6125–6133. doi: 10.1093/nar/gkv519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenstein M., Hipolito C., Lam C., Dietrich D., Perrin D.M. A highly selective DNAzyme sensor for mercuric ions. Angew. Chem., Int. Ed. 2008;47:4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 16.Ono A., Togashi H. Molecular sensors: Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew. Chem., Int. Ed. 2004;43:4300–4302. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]
- 17.Huang P.-J.J., Vazin M., Matuszek Ż., Liu J. A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucleic Acids Res. 2015;43:461–469. doi: 10.1093/nar/gku1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang P.-J.J., Vazin M., Liu J. In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal. Chem. 2014;86:9993–9999. doi: 10.1021/ac5029962. [DOI] [PubMed] [Google Scholar]
- 19.Huang P.-J.J., Lin J., Cao J., Vazin M., Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal. Chem. 2014;86:1816–1821. doi: 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- 20.Huang P.-J.J., Vazin M., Liu J. In vitro selection of a DNAzyme cooperatively binding two lanthanide ions for RNA cleavage. Biochemistry. 2016;55:2518–2525. doi: 10.1021/acs.biochem.6b00132. [DOI] [PubMed] [Google Scholar]
- 21.Ono A., Cao S., Togashi H., Tashiro M., Fujimoto T., Machinami T., Oda S., Miyake Y., Okamoto I., Tanaka Y. Specific interactions between silver(I) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 2008;2008:4825–4827. doi: 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- 22.Saran R., Liu J. A silver DNAzyme. Anal. Chem. 2016;88:4014–4020. doi: 10.1021/acs.analchem.6b00327. [DOI] [PubMed] [Google Scholar]
- 23.Ueyama H., Takagi M., Takenaka S. A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. Fluorescence resonance energy transfer associated with guanine quartet-potassium ion complex formation. J. Am. Chem. Soc. 2002;124:14286–14287. doi: 10.1021/ja026892f. [DOI] [PubMed] [Google Scholar]
- 24.Hoang M., Huang P.-J.J., Liu J. G-quadruplex DNA for fluorescent and colorimetric detection of thallium(I) ACS Sensors. 2015;1:137–143. [Google Scholar]
- 25.Sigel R.K.O., Sigel H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 2010;43:974–984. doi: 10.1021/ar900197y. [DOI] [PubMed] [Google Scholar]
- 26.Torabi S.-F., Wu P., McGhee C.E., Chen L., Hwang K., Zheng N., Cheng J., Lu Y. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W., Saran R., Chen Q., Ding J., Liu J. A new Na+-dependent RNA-cleaving DNAzyme with over 1000-fold rate acceleration by ethanol. ChemBioChem. 2016;17:159–163. doi: 10.1002/cbic.201500603. [DOI] [PubMed] [Google Scholar]
- 28.Geyer C.R., Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 29.Vazin M., Huang P.-J.J., Matuszek Ż., Liu J. Biochemical characterization of a lanthanide-dependent DNAzyme with normal and phosphorothioate-modified substrates. Biochemistry. 2015;54:6132–6138. doi: 10.1021/acs.biochem.5b00691. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W., Zhang Y., Huang P.-J.J., Ding J., Liu J. A DNAzyme requiring two different metal ions at two distinct sites. Nucleic Acids Res. 2016;44:354–363. doi: 10.1093/nar/gkv1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torabi S.-F., Lu Y. Identification of the same Na+-specific DNAzyme motif from two in vitro selections under different conditions. J. Mol. Evol. 2015;81:225–234. doi: 10.1007/s00239-015-9715-7. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W., Ding J., Liu J. A highly selective Na+ aptamer dissected by sensitized Tb3+ luminescence. ChemBioChem. 2016;17:1563–1570. doi: 10.1002/cbic.201600174. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Lu Y. FRETt study of a trifluorophore-labeled DNAzyme. J. Am. Chem. Soc. 2002;124:15208–15216. doi: 10.1021/ja027647z. [DOI] [PubMed] [Google Scholar]
- 34.Kim H.K., Rasnik I., Liu J.W., Ha T.J., Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007;3:762–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y., Lu Y. Metal-ion-dependent folding of a uranyl-specific DNAzyme: Insight into function from fluorescence resonance energy transfer studies. Chem. Eur. J. 2011;17:13732–13742. doi: 10.1002/chem.201100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam J.C.F., Li Y. Influence of cleavage site on global folding of an RNA-cleaving DNAzyme. ChemBioChem. 2010;11:1710–1719. doi: 10.1002/cbic.201000144. [DOI] [PubMed] [Google Scholar]
- 37.Jung J., Han K.Y., Koh H.R., Lee J., Choi Y.M., Kim C., Kim S.K. Effect of single-base mutation on activity and folding of 10-23 deoxyribozyme studied by three-color single-molecule alex fret. J. Phys. Chem. B. 2012;116:3007–3012. doi: 10.1021/jp2117196. [DOI] [PubMed] [Google Scholar]
- 38.Hwang K., Hosseinzadeh P., Lu Y. Biochemical and biophysical understanding of metal ion selectivity of dnazymes. Inorg. Chim. Acta. doi: 10.1016/j.ica.2016.04.017. doi:10.1016/j.ica.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cepeda-Plaza M., Null E.L., Lu Y. Metal ion as both a cofactor and a probe of metal-binding sites in a uranyl-specific DNAzyme: A uranyl photocleavage study. Nucleic Acids Res. 2013;41:9361–9370. doi: 10.1093/nar/gkt694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones A.C., Neely R.K. 2-aminopurine as a fluorescent probe of DNA conformation and the DNA–enzyme interface. Q. Rev. Biophys. 2015;48:244–279. doi: 10.1017/S0033583514000158. [DOI] [PubMed] [Google Scholar]
- 41.Gray R.D., Petraccone L., Trent J.O., Chaires J.B. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soulière M.F., Haller A., Rieder R., Micura R. A powerful approach for the selection of 2-aminopurine substitution sites to investigate RNA folding. J. Am. Chem. Soc. 2011;133:16161–16167. doi: 10.1021/ja2063583. [DOI] [PubMed] [Google Scholar]
- 43.Li M., Sato Y., Nishizawa S., Seino T., Nakamura K., Teramae N. 2-aminopurine-modified abasic-site-containing duplex DNA for highly selective detection of theophylline. J. Am. Chem. Soc. 2009;131:2448–2449. doi: 10.1021/ja8095625. [DOI] [PubMed] [Google Scholar]
- 44.Li N., Ho C.-M. Aptamer-based optical probes with separated molecular recognition and signal transduction modules. J. Am. Chem. Soc. 2008;130:2380–2381. doi: 10.1021/ja076787b. [DOI] [PubMed] [Google Scholar]
- 45.Zhou W., Ding J., Liu J. 2-aminopurine-modified DNA homopolymers for robust and sensitive detection of mercury and silver. Biosens. Bioelectron. 2017;87:171–177. doi: 10.1016/j.bios.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 46.Mazumdar D., Nagraj N., Kim H.-K., Meng X., Brown A.K., Sun Q., Li W., Lu Y. Activity, folding and Z-DNA formation of the 8-17 DNAzyme in the presence of monovalent ions. J. Am. Chem. Soc. 2009;131:5506–5515. doi: 10.1021/ja8082939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law S.M., Eritja R., Goodman M.F., Breslauer K.J. Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry. 1996;35:12329–12337. doi: 10.1021/bi9614545. [DOI] [PubMed] [Google Scholar]
- 49.Hall K.B. 2-Aminopurine as a Probe of RNA Conformational Transitions. Methods Enzymol. 2009;469:269–285. doi: 10.1016/S0076-6879(09)69013-3. [DOI] [PubMed] [Google Scholar]
- 50.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 51.Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown A.K., Li J., Pavot C.M.B., Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 53.Ponce-Salvatierra A., Wawrzyniak-Turek K., Steuerwald U., Höbartner C., Pena V. Crystal structure of a DNA catalyst. Nature. 2016;529:231–234. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- 54.Sigel R.K.O., Pyle A.M. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- 55.Rajendran M., Ellington A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2008;390:1067–1075. doi: 10.1007/s00216-007-1735-8. [DOI] [PubMed] [Google Scholar]
- 56.Qu H., Csordas A.T., Wang J., Oh S.S., Eisenstein M.S., Soh H.T. Rapid and label-free strategy to isolate aptamers for metal ions. ACS Nano. 2016;10:7558–7565. doi: 10.1021/acsnano.6b02558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarkar A.R., Heo C.H., Park M.Y., Lee H.W., Kim H.M. A small molecule two-photon fluorescent probe for intracellular sodium ions. Chem. Commun. 2014;50:1309–1312. doi: 10.1039/c3cc48342h. [DOI] [PubMed] [Google Scholar]
- 58.Kim M.K., Lim C.S., Hong J.T., Han J.H., Jang H.-Y., Kim H.M., Cho B.R. Sodium-ion-selective two-photon fluorescent probe for in vivo imaging. Angew. Chem., Int. Ed. 2010;49:364–367. doi: 10.1002/anie.200904835. [DOI] [PubMed] [Google Scholar]
- 59.Martin V.V., Rothe A., Diwu Z., Gee K.R. Fluorescent sodium ion indicators based on the 1,7-diaza-15-crown-5 system. Bioorg. Med. Chem. Lett. 2004;14:5313–5316. doi: 10.1016/j.bmcl.2004.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.