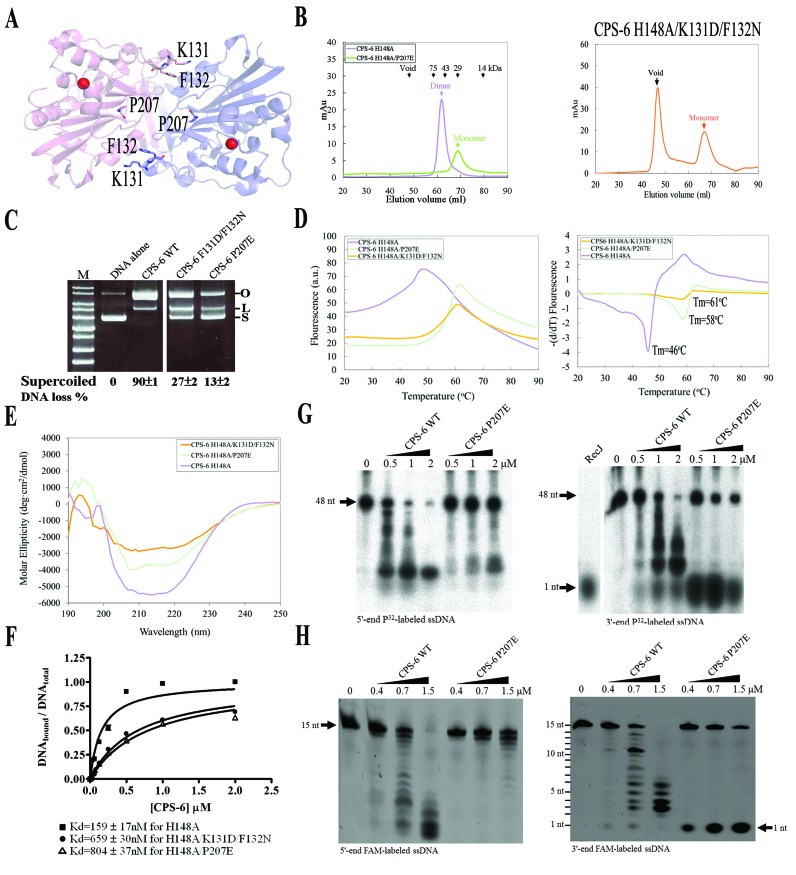
Figure 5.
Dimeric conformation of CPS-6 is critical for its optimal endonuclease activity. (A) P207, K131 and F132 are located in the dimeric interface in CPS-6 dimer based on the crystal structure of CPS-6(H148A) (PDB entry: 3S5B). (B) The CPS-6 H148A/P207E and H148A/K131D/F132N mutants were eluted as monomers in the size exclusion chromatography (Superdex 75), whereas the CPS-6 H148A mutant was eluted as a homodimer. (C) Plasmid-nicking assays show that 90%, 27% and 13% of supercoiled DNA was digested by wild-type CPS-6, K131D/F132N and P207E mutants, respectively. (D) The overall folding stability of CPS-6 mutants was assayed by differential scanning fluorimetry (left panel). The fluorescence intensities for three CPS-6 mutants were normalized for comparison. The melting points were estimated for the three CPS-6 mutant proteins (right panel). (E) Circular dichroism spectra (in molar ellipticity) for CPS-6 H148A, H148A/P207E and H148A/K131D/F132N mutants. (F) Filter binding assays were performed by incubating CPS-6 with a 5′-end P32-labeled single-stranded 48-nucleotide DNA. (G) Wild-type CPS-6 digests the 5′-end P32-labeled single-stranded DNA more efficiently than the P207E mutant (left panel). Wild-type CPS-6 digests the 3′-end P32-labeled single-stranded DNA into small fragments, whereas P207E mainly generated a 3′-end mononucleotide (right panel). (H) Distinct digesting patterns were observed for the wild-type CPS-6 and P207E mutant in digesting 5′-end or 3′-end FAM-labeled single-stranded DNA with a sequence of 5′-AATGTTGGGAGGAAA-3′. P207E digested the 3′-end FAM-labeled single-stranded DNA to yield an instant release of the 3′-end FAM-labeled mononucleotide (right panel).