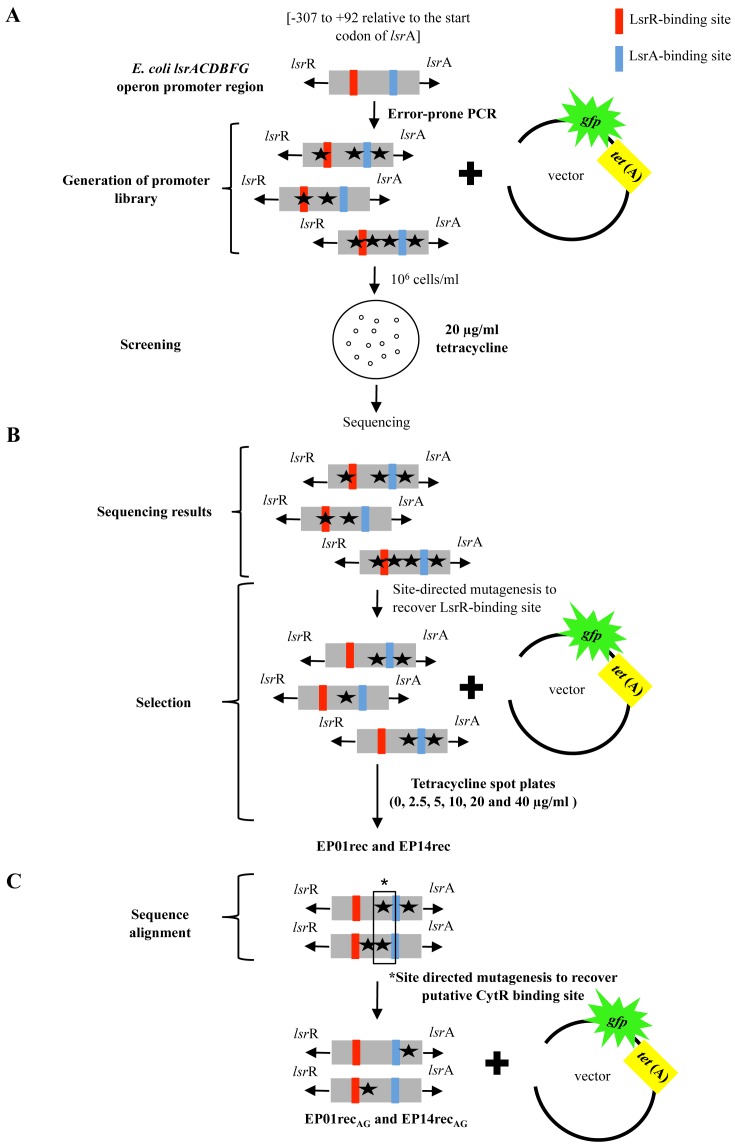
Figure 1.
Schematic of the experimental design used to obtain and to characterize lsr operon promoter mutants from the E. coli lsrACDBFG operon promoter region. (A) Generation of promoter library by ePCR. The strongest promoters were selected based on their ability to confer survival in the presence of 20 μg/ml tetracycline and sequenced to identify the mutations. Surviving clones were denoted as EPxx, where xx refers to the colony number. (B) Identification of mutations on the 14 clones selected against 20 μg/ml tetracycline (EP01-EP14) and site-directed mutagenesis to revert mutations found in the LsrR-binding site region (these clones designated as EPxxrec). These clones were tested for growth at different concentrations of tetracycline (2.5, 5.0, 10, 20 and 40 μg/ml). (C) Sequencing analysis of the two lsr promoters (EP01rec and EP14rec) that maintained the ability to survive in 20 μg/ml tetracycline. Site-directed mutagenesis was used to restore the single mutual nucleotide found to be mutated in both clones, EP01rec and EP14rec, in the putative CytR-binding site sequence (G→A).