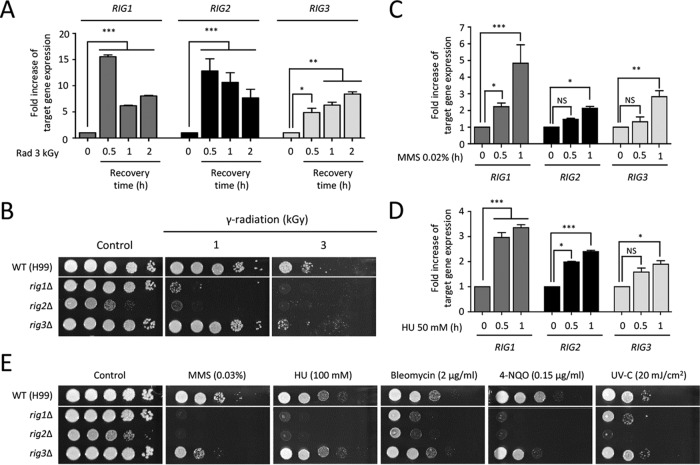
FIG 6 .
Identification of radiation-responsive genes in C. neoformans. (A, C, and D) Quantitative measurement of fold increase in expression of radiation-induced genes. The fold increase of the target gene expression was determined by qRT-PCR analysis with the gene-specific primers listed in Table S2. The cDNA was synthesized with total RNAs extracted from H99 strains recovered 30, 60, and 120 min after exposure to gamma radiation or not exposed to gamma radiation (A). The qRT analysis was performed with the cDNA synthesized from total RNA isolated from WT H99 strains grown in YPD medium containing 0.02% MMS (C) or 50 mM HU (D). Duplicate technical experiments with two independent biological samples were performed. Representative images from independent experiments for each radiation-responsive gene are displayed. Error bars indicate standard deviations. Asterisks indicate statistical significance of differences in the relative expression levels (*, P < 0.05; **, P < 0.01; ***, P < 0.001). NS, not significant. (B and E) RIG1, RIG2, and RIG3 genes were mainly involved in gamma radiation resistance, as well as DNA damage response. Each C. neoformans strain (WT [H99] or rig1Δ [KW158], rig2Δ [KW160], or rig3Δ [KW96] mutant) was grown overnight at 30°C in liquid YPD medium, and 10-fold serially diluted cells (1 to 104 dilutions) were spotted onto the YPD agar medium. Strains were exposed to the indicated dose of gamma radiation for 1 h. For the DNA damage test, 10-fold serially diluted cells were spotted onto YPD agar medium containing the indicated concentration of agents. The two images split by a horizontal white line in each spot assay were obtained from the same plate.