ABSTRACT
This study aimed to understand the exact function and potential mechanism of miR-4500 in colorectal cancer (CRC). In this study, the expression of miR-4500 was decreased in both CRC cells and tissues, and downregulated miR-4500 indicated advanced tumor stage and poor survival. By bisulfite sequencing analysis, we found that the CpG island in the promoter region of miR-4500 was hypermethylated in CRC cells and tissues compared with normal control cells and non-tumor tissues, respectively. Functionally, gain- and loss-of-function analyses indicated the tumor suppressor role of miR-4500: it suppressed cell proliferation, cell cycle progression, migration, and invasion. Predictive algorithms and experimental analyses identified HMGA2 as a direct target of miR-4500. Reintroducing HMGA2 impaired the inhibitory effects of miR-4500 on cell growth and motility. Clinically, higher HMGA2 protein expression in CRC tissues was associated with advanced tumor stage and poor survival. An inverse correlation was found between miR-4500 levels and HMGA2 protein expression. Taken together, this study provides the first evidence that miR-4500 functions as a novel tumor suppressor in the miR-4500/HMGA2 axis in colorectal carcinogenesis, and restoring miR-4500 expression might represent a promising therapeutic strategy for CRC.
KEYWORDS: Colorectal cancer, cell proliferation, HMGA2, invasion, miR-4500, methylation, prognosis
Abbreviations
- CRC
Colorectal cancer
- HMGA2
high mobility group AT-hook 2
- 5-aza
5-aza-2′-deoxycytidine
Introduction
Colorectal cancer (CRC) is one of the most common carcinomas worldwide, and the 5-year survival rate of CRC patients ranges from 40% to 60%.1 Metastasis is the main obstacle for effective CRC therapy. Understanding the key regulators in colorectal carcinogenesis is indispensable for identifying novel, improved anticancer strategies.
MicroRNAs (miRNAs) are small non-coding RNAs (18–25 nucleotides) that modulate gene expression via sequence-specific base pairing on the 3′-UTR of target mRNA.2 An increasing number of studies have demonstrated that miRNAs play critical roles in several tumor-related biological processes, including cell proliferation, apoptosis, migration, and invasion.3,4 Dysregulated miRNA expression is involved in the development of cancer, and miRNAs can be used as diagnostic or prognostic biomarkers.5 Thus, exploring the novel regulators of tumorigenesis may have benefits for early diagnostics and for understanding the underlying molecular mechanism. The sequence of miR-4500 was previously determined,6,7 and the suppressive effect of miR-4500 on tumor cell growth was first reported in non-small cell lung cancer (NSCLC).8 MiR-4500 shares a seed sequence with the let-7 family and miR-98-5p, which are frequently described as tumor suppressors that inhibit cell proliferation, migration, and angiogenesis. Furthermore, using the CAGE database, we found that the expression of miR-4500 host gene (MIR4500HG)-related transcripts (AF339814) was significantly different between normal and malignant liver samples. We speculated that miR-4500/MIR4500HG may play a role in tumorigenesis. However, the exact role of miR-4500 in CRC progression was unclear.
High Mobility Group AT-2 hook (HMGA2) was first reported as an oncogene,9 and this protein is frequently upregulated in several cancers, including CRC.10 Overexpression of HMGA2 in Rat 1a cells enables tumor growth in nude mice.11 The stimulatory effect of HMGA2 on tumor cell behavior has been observed in many types of cancer.12-14 Accumulating evidence shows that upregulated HMGA2 is indicative of tumor progression and poor survival.10,15-17 In addition, HMGA2 is involved in miRNA-regulated cancer progression.18,19 However, whether other miRNAs are involved in modulating HMGA2 in CRC carcinogenesis is unknown.
In this study, we first confirmed that miR-4500 expression levels were epigenetically downregulated in CRC. Then, our results showed that miR-4500 inhibited CRC cell proliferation, migration, and invasion in vitro and tumor growth in vivo. The tumor inhibitory effect of miR-4500 could be partially reversed by overexpression of HMGA2, a direct target of miR-4500. MiR-4500 expression was negatively correlated with HMGA2 expression in clinical samples. These data provide novel insights into the potential mechanism of CRC tumorigenesis.
Materials and methods
Clinical samples and cell lines
A total of 75 pairs of CRC tissues and adjacent non-tumor tissues were collected from patients in Zhanjiang People's Central Hospital between Feb 2009 and Jul 2010. This study was approved by the Medical Ethics Committee of the hospital and written informed consent were obtained from patients. None of the patients received chemo- or radiotherapy before surgery. The clinicopathological features were obtained from medical records. Corresponding adjacent normal tissues were obtained 2 cm beyond the boundary of CRC tissues. All primary tumors and corresponding normal tissues were recognized by 2 junior pathologists, followed by one senior pathologist. Both snap-frozen tissue samples (stored at −80°C) and Formalin-fixed and paraffin-embedded (FFPE) specimens were used in this study. Human CRC cell lines (HT-29, LoVo, HCT-116, and SW480) were cultured at 37°C and 5% CO2 in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen). A normal colon epithelial cell line (FHC) was grown in DMEM:F12 medium and used as control cells.
Quantitative real-time PCR (RT-qPCR) and demethylation agent treatment
MiRNAs were isolated from tissue specimens and cells using RNAiso kit (Takara, China) and reversely transcribed into cDNA using One Step PrimeScript miRNA cDNA Synthesis Kit (Takara). The resulting cDNA were quantified using an ABI 7500 PCR System (ABI, Foster City, CA, USA). Small nuclear RNA U6 was used as internal control and expression level of miR-4500 was calculated using 2−ΔΔCt method. For demethylation analysis, HCT-116 and SW480 cells were cultured for 72 h with 0, 2.5 µM and 5µM 5-aza-2′-deoxycytidine (5-aza; Sigma, St Louis, MO, USA) with medium changed every 24 h.
Identification of the miR-4500 promoter and methylation analysis
The potential transcriptional start site (TSS) and promoter for miR-4500/MIR4500HG were identified using the CAGE database (http://fantom3.gsc.riken.jp) and FPROM (http://molbiol-tools.ca/Promoters.htm). Four candidate promoter/enhancer sites (position 1, chr13: 88332642; position 2, 88330780; position 3, 88329738; and position 4, 88330145) were reported by FPROM. Of these 4 promoters, only position 1 was close to the potential TSS of miR-4500, which was identified using CAGE tag data. Luciferase activity assays were performed to identify the promoter activity of this region (∼0.6 kb upstream of the gene). Briefly, promoter of miR-4500 was amplified by PCR using a BAC clone as a template with forward: 5′-ttacggctgcggacta-3′ and reverse: 5′-tcgagattcgggtcaat-3′ primers. Products were then cloned into a pGL3-Basic firefly luciferase vector. Then, 293T cells grown in 6-well plates were cotransfected with 2 μg of the firefly luciferase vector containing promoter region and 10 ng of CMV-renilla luciferase vector (Promega, Madison, WI, USA) using Dharmafect Duo transfection reagent (Dharmacon, Lafayette, CO).
Bisulfite sequencing-PCR (BSP) analysis was conducted to evaluate the methylation status of miR-4500 promoter (from −641 to −303). DNA isolated from cell lines and tissues was modified using EpiTect Bisulfite Kit (Qiagen, Germany) and primers for bisulfite sequencing were designed using Methyl Primer Software Version 1.0 (ABI) as follows: forward: 5′-tatgtttgtggtttgttttgta-3′; reverse: 5′-aatccaaaactctctctcttcc-3′. PCR-amplified product was transformed into E.coli cells and the resulting plasmids were subjected to sequencing.
Oligonucleotide transfection, plasmid construction, and Luciferase reporter gene assays
The miR-4500 mimics, miR-4500 inhibitor, mimic negative control (NC-mim), and inhibitor negative control (NC-inh) were obtained from GenePharma (Shanghai, China) and transfected into cells using Lipofectamine 2000 (Invitrogen). HMGA2 cDNA without its 3′-UTR was inserted into pcDNA3.1(+) to rescue the expression of HMGA2 in mimic-treated cells. The sequence containing miR-4500 pre-miRNA was amplified by PCR from genomic DNA using the following primers: forward: 5′-cttgaacagtgctttcaggccaaac-3′; and reverse: 5′-tctagaatcacgcacatgagaatt-3′. The PCR products were cloned into the BamH/Xho I sites of pcDNA 3.1(+). HCT-116 cells at 70% confluency were transfected with the pcDNA3.1-miR-4500 expression vector or an empty vector (mock), which served as a control. Stable cells were selected with G418 (400 mg/ml), and after 2 weeks, single-cell clones were selected and expanded.
To construct the luciferase reporter plasmids, the full-length 3′-UTR of HMGA2 was sub-cloned into pMIR-REPORT Luciferase (Invitrogen) to generate HMGA2 3′-UTR-WT. HMGA2-3′-UTR-Mut was created using a Phusion mutagenesis kit (NEB, MA, USA). The following primers were used: sense: 5′-caaagacctacctccagacttcaaaagg-3′; ntisense: 5′-ccttttgaagtctggaggtaggtctttg-3′ for wide type. The primers: sense: 5′- caaagacgatcctccagacttcaaaagg −3′; antisense: 5′- ccttttgaagtctggaggatcgtctttg −3′ for mutant type. Cells were co-transfected with 1 µg of wide-type or mutants plasmid and 50 ng of Renilla luciferase plasmid. 24 h later, cells were lysed and luciferase activity was measured with the Dual-Luciferase reporter assay system (Promega).
Cell proliferation and cell cycle analysis
Cell proliferation was measured using the MTT assay. Cells (1.5 × 103 ) were seeded into 96-well plates, and 20 µl MTT (5 mg/ml; Sigma) was added to each well at 0, 24, 48, and 72 h. Cell viability was measured by measurement of absorbance at 570 nm. For cell cycle assay, cells were fixed 70% ethanol at 4°C overnight. Then the fixed cells were treated with RNase A (0.1 mg/ml) and propidium iodide (PI, 0.05 mg/ml) at 37°C for 30 min, and then analyzed by flow cytometry (FACSCalibur, Beford, MA, USA).
Wound healing and matrigel transwell invasion assay
Cells were seeded into 6-well plates and grown to 90% confluence. Monolayers in the center of the wells were scraped using a 200 µl pipette tips. Cell movement into the wound area was photographed at 0 and 24 h. For the transwell invasion assay, cells suspended in DMEM without FBS were placed on the top chamber of each insert (Millipore, MA, USA) pecoated with 40 µl matrigel (1 mg/ml). After 24 h, the invasive cells crossed to the lower surface were fixed and stained.
Tumor xenografts in vivo
Animal experimental procedures were approved by the Institutional Animal Care and Use Committee of the Guangdong Medical University. HCT-116 cells transfected with stably-expressing miR-4500 (5 × 106 cells/150 µl) vector were injected subcutaneously into the left flanks of nude mice and the right flanks were injected with control vector (mock). All mice (n = 6) were sacrificed 4 weeks later and tumor weight were determined. Tumor samples were excised for Immunohistochemistry (IHC) assay.
IHC analysis
All FFPE samples were cut into 4 µm-thick sections and subjected to standard procedures to detect HMGA2 protein. The tissue sections from xenograft tumors were analyzed for Ki-67 and HMGA2 expression. IHC scoring was based on 2 variables: the proportion of positive cells (0%, 0; 1–50%, 1; 51–75%, 2; and ≥76 %, 3) and the staining intensity (negative-weak, 1; moderate, 2; and strong, 3). The sum of the proportion score and the intensity score was calculated to obtain a total score that ranged from 1 to 6. A score of <4 was defined as low expression, and a score of ≥4 was defined as high expression.
Western blot analysis
Equivalent proteins from each samples were separated by SDS-PAGE and then transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). Membranes were blocked overnight in 5% free-fat milk and incubated for 2 h with primary anti-HMGA2 antibody (ab97276,1:1000; Abcam, Cambridge, MA, USA) or GAPDH (ab37168, 1:2000; Abcam) overnight at 4°C. After 3 washes, membranes were incubated using horseradish peroxidase-linked goat anti-mouse or goat anti-rabbit IgG antibodies and visualized using SuperSignal system (Thermo, Rockford, IL, USA).
Statistical analysis
Data are presented as the mean ± SEM (standard error of the mean) of at least 3 independent experiments after analysis with SPSS 19.0 (IBM, Chicago, IL, USA) or Prism 5.0 (Graph Pad Software, La Jolla, CA, USA). Student's t-test was used to contrast quantitative data between 2 test groups, while 2-way ANOVA was used to compare the differences in quantitative data among 3 or more groups. Survival estimates were calculated using the Kaplan-Meier method and compared using the log-rank test. X2 or Fisher's exact test were used to evaluate the correlations between clinicopathological features and the expression of miR-4500 and HMGA2. Cox proportional hazards regression models were applied in univariate and multivariate analyses. Spearman's correlation was used to identify the correlation between miR-4500 levels and HMGA2 protein expression. P < 0.05 was considered to indicate significance.
Results
MiR-4500 was downregulated in CRC and correlated with tumor progression and overall survival
To ascertain the endogenous level of miR-4500, we first detected miR-4500 expression levels in CRC cell lines. As shown in Fig. 1A, the expression of miR-4500 was significantly decreased in CRC cells compared to FHC cells. The expression of miR-4500 was reduced in primary CRC tissues compared with adjacent non-tumor tissues (Fig. 1B, p = 0.0073). We separated CRC cases into 2 groups according to the median level of miR-4500 expression in tumors (low group, ≤median; high group, >median) and found that low miR-4500 expression was correlated with larger tumor size (p = 0.025), high TNM stage (p = 0.051), tumor depth (p = 0.014) and lymph node metastasis (p = 0.028; Table 1).
Figure 1.
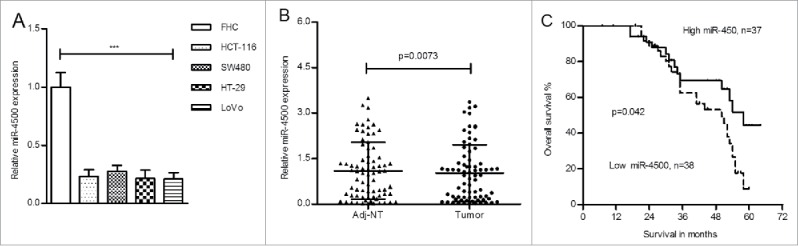
MiR-4500 was downregulated in CRC cells and tissues. (A) The expression levels of miR-4500 were significantly reduced in CRC cells compared with FHC cells. Each bar represents the mean ± SD of 3 independent experiments. (B) The levels of miR-4500 were lower in tumors than in normal tissues; the data are presented as the mean ± SD (paired-samples t test). (C) Downregulated miR-4500 in CRC patients predicted poor overall survival. U6 was used as a loading control. ***P < 0.001.
Table 1.
Correlations between the level of miR-4500 and HMHA2 protein and clinicopathological features of CRC patients.
| Total | MiR-4500 expression |
HMGA2 protein |
|||||
|---|---|---|---|---|---|---|---|
| Variables | n = 75 | high, n = 37 | low, n = 38 | p | high, n = 39 | low, n = 36 | p |
| Gender | 0.520 | 0.217 | |||||
| Female | 24 | 11 | 13 | 15 | 9 | ||
| Male | 51 | 26 | 25 | 24 | 27 | ||
| Age (years) | 0.908 | 0.307 | |||||
| <60 | 37 | 18 | 19 | 17 | 20 | ||
| ≥60 | 38 | 19 | 19 | 22 | 16 | ||
| Differentiation | 0.138 | 0.170 | |||||
| Well, moderate | 46 | 23 | 23 | 21 | 25 | ||
| Poorly | 29 | 14 | 15 | 18 | 11 | ||
| Tumor size (cm) | 0.025 | 0.120 | |||||
| <4 | 43 | 26 | 17 | 19 | 24 | ||
| ≥4 | 32 | 11 | 21 | 20 | 12 | ||
| TNM stage | 0.051 | 0.008 | |||||
| I, II | 36 | 22 | 14 | 13 | 23 | ||
| III, IV | 39 | 15 | 24 | 26 | 13 | ||
| T stage | 0.014 | 0.200 | |||||
| T1–2 | 40 | 25 | 15 | 18 | 22 | ||
| T3–4 | 35 | 12 | 23 | 21 | 14 | ||
| N stage | 0.028 | 0.004 | |||||
| N0 | 37 | 23 | 14 | 13 | 24 | ||
| N1–2 | 38 | 14 | 24 | 26 | 12 | ||
| Location | 0.430 | 0.595 | |||||
| Colon | 42 | 19 | 23 | 23 | 19 | ||
| Rectum | 33 | 18 | 15 | 16 | 17 | ||
The Kaplan-Meier analysis showed that downregulated miR-4500 was correlated with poor overall survival (Fig. 1C, p = 0.042). To exclude the confounder effect, we also performed Cox proportional hazards regression analysis. The univariate analysis showed that low miR-4500 levels, age (>60 years), differentiation (poor), higher TNM stage (III/IV), and lymph node metastasis were significant risk factor for overall survival, whereas only TNM stage [p = 0.025, hazard ratio (HR) = 2.268, 95% confidence interval (CI) = 1.106–4.651] and differentiation (p = 0.019, HR = 2.187, 95% CI = 1.135–4.215) were confirmed as independent risk factors in the multivariate analysis (Table 2).
Table 2.
Univariate and multivariate Cox's hazards model analysis for prognostic factors.
| Univariate |
Multivariate |
|||
|---|---|---|---|---|
| Variables | HR (95% CI) | p | HR (95% CI) | p |
| Gender (female/male) | 1.139 (0.576–2.252) | NS | ||
| Age (<60 y/≥60 y) | 1.903 (1.010–3.588) | 0.047 | 1.537 (0.776–3.042) | NS |
| Differentiation (well, mod/poorly) | 2.611 (1.373–4.964) | 0.003 | 2.187 (1.135–4.215) | 0.019 |
| Tumor size (<4 cm/≥4 cm) | 0.952 (0.506–1.791) | NS | ||
| TNM stage (I, II/III, IV) | 2.712 (1.342–5.479) | 0.005 | 2.268 (1.106–4.651) | 0.025 |
| T stage (T1−2/T3–4) | 1.275 (0.674–2.412) | NS | ||
| N stage (N0/N1–2) | 2.455 (1.237–4.870) | 0.010 | 1.899 (0.997–3.476) | NS |
| Location (colon/rectum) | 1.682 (0.797–3.255) | NS | ||
| MiR-4500 (high/low) | 1.961 (0.999–3.850) | 0.050 | 1.774 (0.880–3.578) | NS |
| HMGA2 (low/high) | 2.113 (1.082–4.127) | 0.028 | 1.481 (0.725–3.029) | NS |
Mod, moderate; HR, hazard ratio; CI, confidence interval; NS, none significance.
Mir-4500 is an epigenetic target in CRC
We were interested in the potential mechanism by which miR-4500 is downregulated in CRC cells and tissues. Using the UCSC genome browser (February 2009), we determined that miR-4500 is located within the introns of MIR4500HG (Fig. 2A). One region upstream of MIR4500HG (∼0.6 kb) was identified as a potential TSS for miR-4500 based on CAGE tag data. This region had 4 CAGE tags. Using the UCSC genome browser, we found a marker of histone modification (H3K4me1) in this region, as well as an active marker of histone modification (H3K4me3), suggesting the possibility of epigenetic modification in this region (Fig. S1). We cloned the promoter region into the pGL3-basic reporter vector and found that this region showed strong promoter activity compared to vector control (Fig. S2), suggesting promoter activity in this region (∼0.6 kb upstream of the gene).
Figure 2.
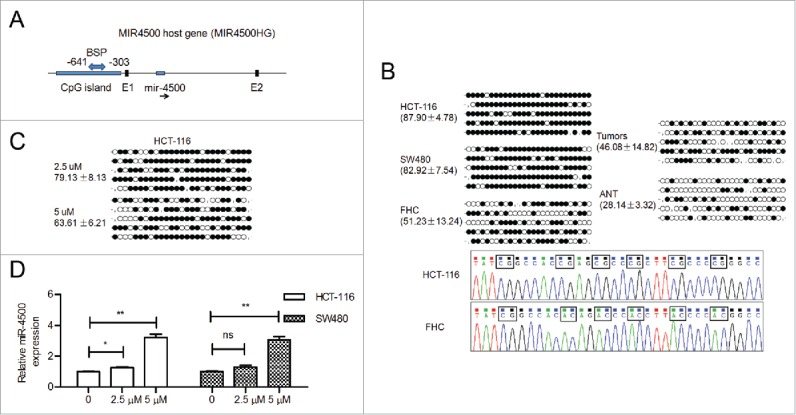
MiR-4500 was epigenetically downregulated in CRC. (A) MiR-4500 is embedded in the intron located between the first (E1) and second exons (E2) of MIR4500HG. A CpG island is located in the promoter region of MIR4500HG. (B) Representative bisulfite sequencing results of the methylation status of the CpG island in cells and tissues; ANT, adjacent non-tumor tissues. The circles indicate unmethylated (open circles) and methylated (solid circles) CpG sites. The percentage of methylated CpG sites is presented as the mean ± SD. (C) Representative BSP results of HCT-116 cells after 5-aza treatment. (D) The expression levels of miR-4500 in 5-aza-treated cells. The data are presented as the mean ± SEM. ns, not significant; *P < 0.05; **P < 0.01.
Bisulfite sequencing was conducted in CRC cells and in paired tumor and adjacent non-malignant tissues to determine the CpG methylation status of the miR-4500/MIR4500HG promoter. Representative bisulfite sequencing data on 27 CpG sites within a 339-bp promoter region of miR-4500 are shown. The percentage of methylated CpG sites was higher in CRC cells and primary tumors compared with FHC cells and adjacent non-tumor tissue, respectively (Fig. 2B). Subsequently, the CpG islands were demethylated by 5-aza treatment of HCT-116 cells (Fig. 2C). All of the 5-aza-treated cells, except those treated with 2.5 µM 5-aza, showed obviously increased miR-4500 expression compared with control cells (Fig. 2D). These findings suggested that DNA methylation may be involved in silencing miR-4500 during the development of CRC.
MiR-4500 inhibited tumor cell growth and motility
SW480 cells were used in the knockdown analysis because of their relatively high expression of miR-4500. HCT-116 cells were utilized to overexpress miR-4500 because they are easier to transfect, they are tumorigenic, and they are derived from a carcinoma, all of which make them different from other cell lines. The diversity of CRC cells may prove the prevalence of aberrant miR-4500 expression. We transfected HCT-116 and SW480 cells with mimics and inhibitor, respectively; miR-4500 expression increased in HCT-116 cells by 11.62-fold and decreased in SW480 cells by 3.57-fold compared to control cells (Fig. 3A). Overexpression of miR-4500 inhibited the proliferation of HCT-116 cells, while downregulation of miR-4500 accelerated the proliferation of SW480 cells (Fig. 3B). Flow cytometry assays indicated that overexpressing miR-4500 in HCT-116 cells led to a significant increase in the population of cells in G0/G1 phase and a decreased percentage of those in S phase, whereas inhibiting miR-4500 expression markedly increased the S phase cell population (Fig. 3C). Therefore, the suppression of cell cycle progression at the G1/S transition may be an explanation for the growth-inhibitory role of miR-4500 in CRC cells.
Figure 3.
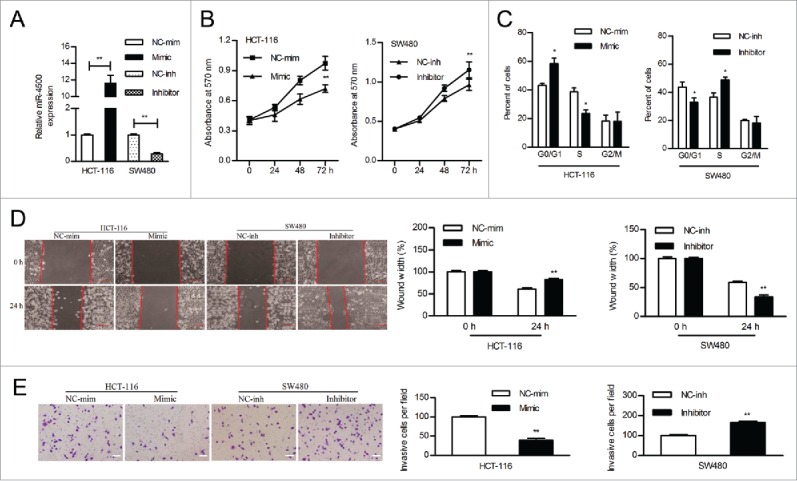
MiR-4500 inhibited cell growth and motility in vitro. (A) The expression level of miR-4500 in CRC cells transfected with mimics or inhibitor. (B) The effects of overexpressing or inhibiting miR-4500 on CRC cell proliferation were determined by MTT assay. (C) The cell cycle distribution of CRC cells was assayed by flow cytometry. Wound healing (D) and transwell invasion (E) assays were performed to evaluate the effect of miR-4500 on cell migration and invasion, respectively; scale bar: 100 µm. Each bar represents the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01.
CRC cell migration was demonstrated in wound healing assays. HCT-116 cells treated with miR-4500 mimics were noticeably less migratory than those treated with the mimic control, whereas SW480 cells treated with the inhibitor showed increased migration (Fig. 3D). Matrigel Transwell assays showed that miR-4500 significantly inhibited HCT-116 cell invasion, while the suppression of miR-4500 enhanced SW480 cell invasion (Fig. 3E).
HMGA2 was a direct target of miR-4500
HMGA2 was one of the putative target genes that were predicted by 3 publically available databases (TargetScan, miRDB, and microRNA). We focused on HMGA2 due to a previous study that reported increased HMGA2 levels in CRC.10 In addition, using a publically available database (www.oncomine.org), we found that HMGA2 mRNA levels were significantly increased in colorectal adenocarcinoma in the TCGA dataset (p = 1.27 × 10−6, fold change = 1.891). We identified potential miR-4500 binding sites in the highly conserved 3′-UTR of HMGA2 mRNA (Fig. 4A). Overexpression of miR-4500 significantly reduced the luciferase activity of wild-type HMGA2 3′-UTR but not of a mutant HMGA2 3′-UTR in CRC cells (Fig. 4B). Upregulation of miR-4500 significantly reduced HMGA2 mRNA and protein levels in both HCT-116 and SW480 cells (Fig. 4C and D). Our data demonstrated that HMGA2 was a direct target of miR-4500.
Figure 4.
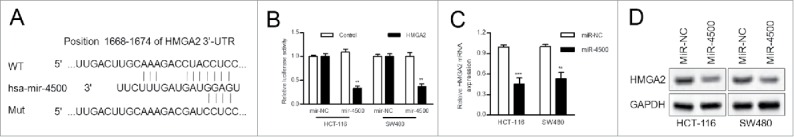
MiR-4500 directly targets the HMGA2 3′-UTR. (A) The putative miR-4500 binding sites in the 3′-UTR of HMGA2 mRNA. (B) A dual luciferase report assay was performed. Cells were co-transfected with wild-type or mutant reporters and the negative control or miR-4500. (C) The mRNA levels of HMGA2 were determined by RT-qPCR after transfection of mimics. (D) Protein expression of HMGA2 in cells that were stably transfected with miR-4500. Bars indicate the normalized mean fluorescence intensity ± SD for 3 independent experiments. **P < 0.01.
HMGA2 regulated miR-4500-mediated tumor behavior in vitro and in vivo
To further investigate whether HMGA2 was involved in the tumor suppressive effects of miR-4500 on CRC cells, we rescued HMGA2 expression in CRC cells (Fig. 5A and B). MTT assays showed that reintroducing HMGA2 significantly impaired miR-4500-mediated inhibition of cell proliferation (Fig. 5C) and cell cycle progression (Fig. 5D). Ectopic HMGA2 expression promoted the migration (Fig. 5E) and invasion (Fig. 5F) of HCT-116 cells transfected with miR-4500 mimics. These in vitro results indicated that HMGA2 was a functional regulator of miR-4500 in CRC.
Figure 5.
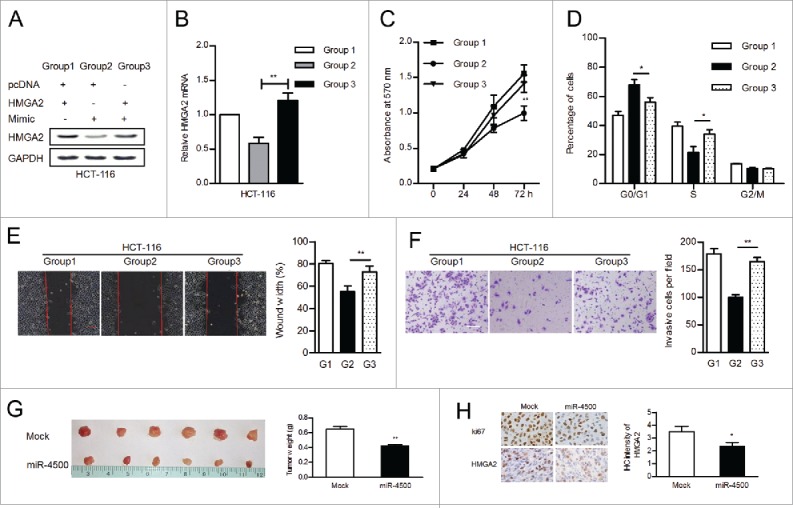
HMGA2 was involved in miR-4500-regulated tumor growth in vitro and in vivo. (A) The expression of HMGA2 was examined by western blot. (B) The mRNA levels of HMGA2 were confirmed by RT-qPCR. The reintroduction of HMGA2 promoted cell growth and motility, as determined by MTT (B), flow cytometry (C), wound healing (D), and transwell (E) assays; scale bar: 100 µm. (F) HCT-116 cells (transfected with miR-4500 or mock) were subcutaneously injected into nude mice; tumors were harvested and weighed. (G) The expression of Ki-67 and HMGA2 in xenograft tumors was evaluated by IHC. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01.
The stable expression of miR-4500 in HCT-116 cells inhibited tumor formation and significantly decreased tumor weight compared with the control vector (Fig. 5G). Then, we performed IHC on xenograft tumors and found that HMGA2 protein levels were obviously reduced in miR-4500-overexpressing tumors compared with control tumors (Fig. 5H).
HMGA2 was negatively correlated with miR-4500 levels in CRC tissues
The correlations between miR-4500 and HMGA2 were identified based on in vitro and in vivo analyses. Therefore, we aimed to determine whether HMGA2 is regulated by miR-4500 in CRC tissues. We assessed the protein levels of HMGA2 in tumor tissues by IHC (Fig. 6A). We separated primary tumor samples into 2 groups according to the HMGA2 staining score and found that high HMGA2 levels significantly correlated with higher TNM stage and lymph node metastasis (p = 0.008 and p = 0.004, respectively, Table 1). Patients with low HMGA2 protein expression had a better prognosis compared to those with high HMGA2 protein expression (Fig. 6B, p = 0.022). The prognostic significance of HMGA2 was confirmed by univariate analysis, while the multivariate analysis showed that HMGA2 was not an independent risk factor. As expected, the expression levels of miR-4500 were inversely correlated with HMGA2 protein expression (Table 3). These data suggested that HMGA2 is a prognostic factor and a functional target of miR-4500 in clinical cases.
Figure 6.
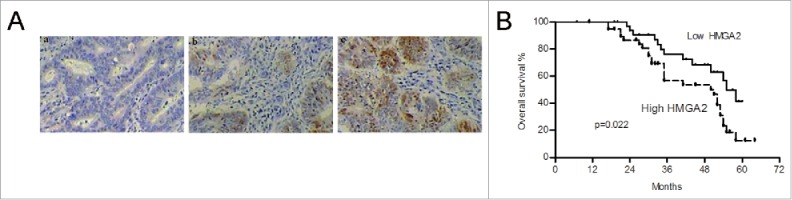
The protein levels of HMGA2 were correlated with prognosis and inversely associated with miR-4500. (A) IHC of HMGA2 protein in CRC tissues. (a-c) Magnification, × 200. (B) Low protein levels of HMGA2 predicted better survival.
Table 3.
Association of the miR-4500 levels with the protein expression of HMGA2.
| MiR-4500 level | |
|---|---|
| Low HMGA2 expression | 0.122 ± 0.095 |
| High HMGA2 expression | 0.085 ± 0.088 |
| Spearman's correlation, r = −0.228,p = 0.049 |
Discussion
The application of miRNAs for the diagnosis and treatment of cancer is attracting increasing attention due to accumulating evidence of their frequent deregulation in many types of cancer, including CRC.5 In this study, miR-4500 significantly inhibited cell growth and motility and was epigenetically downregulated in CRC. We also identified a direct and functional target of miR-4500, HMGA2, whose expression levels inversely correlated with those of miR-4500 in CRC patients.
In the present study, we used gain- and loss-of-function approaches to ascertain the biological effect of miR-4500 on CRC cell behavior. Our data indicated that miR-4500 suppressed CRC cell proliferation, cell cycle progression, migration and invasion. A previous study also reported that miR-4500 overexpression inhibited NSCLC proliferation and induced apoptosis.8 Our study extended the knowledge of the biological function of miR-4500 to CRC and suggested that miR-4500 is a tumor suppressor. Deregulated miR-4500 may be used as a prognostic factor. More interestingly, we showed a potential mechanism for the downregulation of miR-4500: the CpG island in the MIR4500HG promoter was hypermethylated in CRC cells and primary tumors. Accumulating evidence indicates that several tumor suppressor miRNAs are epigenetically silenced by promoter DNA methylation in cancer.20,21 Thus, restoring miR-4500 expression may be a novel therapeutic strategy for CRC.
By using publically available databases and experimental analysis, HMGA2 was identified as a direct target of miR-4500. Restoring HMGA2 expression attenuated the tumor-suppressive effects of miR-4500 on CRC cells. These data demonstrated that HMGA2 is a functional regulator of the effects of miR-4500 on tumor cell viability and motility. In NSCLC, miR-4500 directly targets LIN28B, which has been consistently reported to be associated with let-7 and miR-98.22,23 TargetScan, miRDB, and TargetMiner revealed that genes other than LIN28B, such as CDC34, PBX3, and FZD3, were predicted as candidate miR-4500 targets. PBX3 is overexpressed in gastric cancer and prostate cancer 24,25 and promotes CRC migration by regulating the MAPK/ERK pathway.26 CDC34 is highly expressed in multiple myeloma (MM) and associated with the growth and survival of MM cells.27 An immunocytochemical study showed that FZD3 is strongly associated with CRC progression.28 Additional studies on miR-4500 and its target genes may be helpful for understanding tumorigenesis. Collectively, similar to the let-7 family, the potential tumor suppressor miR-4500 may affect tumorigenesis by targeting many critical oncogenes.
HMGA2, which is upregulated in many malignancies, is simultaneously regulated by many miRNAs, including miR-204, miR-145, miR-154, and let-7.18,19,29,30 Our findings defined the oncogenic role of HMGA2 in CRC. High expression of HMGA2 was correlated with advanced tumor stage and lymph node metastasis, which was consistent with previous studies.16,31,32 Several previous studies reported that HMGA2 is a metastasis-related protein in cancer;32,33 these findings were confirmed in our study, which showed that HMGA2 overexpression enhanced CRC cell migratory and invasive behavior. Although the multivariate analysis indicated that HMGA2 was not an independent prognostic factor, both the Kaplan-Meier and univariate analyses confirmed the prognostic significance of HMGA2. Taken together, HMGA2 was a key regulator of the effects of miR-4500 on colorectal carcinogenesis.
Previous studies suggested that the let-7 family regulates critical development-related genes. Let-7 family members function in combination to affect both early and late developmental timing decisions34 and are involved in organ development by regulating TGFβ-R1 and E2F5.35,36 Mlin41 is a let-7 target gene in mouse neural tube closure, suggesting that Hlin41 might be a potential human development gene.37 HMGA2 is an effector that is involved in the genetic switch wherein let-7 activation in the fetal state causes a shift toward an adult-like myeloid-dominant output.38 MiR-98, which belongs to the let-7 family, is overexpressed in fetal sclera compared to adult sclera.39 Considering that miR-4500 shares a seed sequence with let-7/miR-98, we speculated that miR-4500 may have functional role in human development.
This study is the first to identify miR-4500 as a novel tumor suppressor and regulator of HMGA2. With our in vitro and in vivo studies, we established the function of miR-4500 and HMGA2 in cell growth, migration, and invasion. Both miR-4500 and HMGA2 have promising prognostic value in CRC. Taken together, our findings highlight that miR-4500 functions as a tumor suppressor and might represent a promising therapeutic target for CRC.
Supplementary Material
Disclosure of potential conflicts of interest
The authors declared that they have no potential conflicts of interest.
Funding
This study was supported by National Natural Science Foundation of China (81173240), Guangdong Major Scientific and Technological Special Project (2013A02210003), Natural Science Foundation of Guangdong Province (2014A030307001), Science and Technology Fund of Zhanjiang (2014A01014), Fund of Guangdong Medical College (Z2013001 and B2013019).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2004; 63:11-30; PMID:23335087; http://dx.doi.org/14744438 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Shi ZM, Jiang CF, Liu X, Chen QD, Qian X, Li DM, Ge X, Wang XF, Liu LZ, et al.. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget 2014; 5:5416-27; PMID:24980823; http://dx.doi.org/ 10.18632/oncotarget.2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farhana L, Dawson MI, Fontana JA. Down regulation of miR-202 modulates Mxd1 and Sin3A repressor complexes to induce apoptosis of pancreatic cancer cells. Cancer Biol Ther 2015; 16:115-24; PMID:25611699; http://dx.doi.org/ 10.4161/15384047.2014.987070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 2010; 79:313-20; PMID:21412018; http://dx.doi.org/ 10.1159/000323283 [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006; 34:D140-4; PMID:16381832; http://dx.doi.org/ 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jima DD, Zhang J, Jacobs C, Richards KL, Dunphy CH, Choi WW, Au WY, Srivastava G, Czader MB, Rizzieri DA, et al.. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood 2010; 116:e118-27; PMID:20733160; http://dx.doi.org/ 10.1182/blood-2010-05-285403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Qian J, Qiang Y, Huang H, Wang C, Li D, Xu B. Down-regulation of miR-4500 promoted non-small cell lung cancer growth. Cell Physiol Biochem 2014; 34:1166-74; PMID:25277326; http://dx.doi.org/ 10.1159/000366329 [DOI] [PubMed] [Google Scholar]
- 9.Manfioletti G, Giancotti V, Bandiera A, Buratti E, Sautière P, Cary P, Crane-Robinson C, Coles B, Goodwin GH. cDNA cloning of the HMGI-C phosphoprotein, a nuclear protein associated with neoplastic and undifferentiated phenotypes. Nucleic Acids Res 1991; 19:6793-7; PMID:1762909; http://dx.doi.org/ 10.1093/nar/19.24.6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, Hu S, Yao L, Peng J, Loera S, et al.. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res 2011; 17:2570-80; PMID:21252160; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood LJ, Maher JF, Bunton TE, Resar LM. The oncogenic properties of the HMG-I gene family. Cancer Res 2000; 60:4256-61; PMID:10945639 [PubMed] [Google Scholar]
- 12.Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, Liang L, Hu J, Shen H, Chen Z, et al.. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int J Oncol 2014; 44:2050-8; PMID:24682444; http://dx.doi.org/ 10.3892/ijo.2014.2360 [DOI] [PubMed] [Google Scholar]
- 13.Malek A, Bakhidze E, Noske A, Sers C, Aigner A, Schäfer R, Tchernitsa O. HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int J Cancer 2008; 23:348-56; http://dx.doi.org/ 10.1002/ijc.23491 [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z, You T. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett 2014; 355:130-40; PMID:25218351; http://dx.doi.org/ 10.1016/j.canlet.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 15.Yang GL, Zhang LH, Bo JJ, Hou KL, Cai X, Chen YY, Li H, Liu DM, Huang YR. Overexpression of HMGA2 in bladder cancer and its association with clinicopathologic features and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur J Surg Oncol 2011; 37:265-71; PMID:21273026; http://dx.doi.org/ 10.1016/j.ejso.2011.01.004 [DOI] [PubMed] [Google Scholar]
- 16.Piscuoglio S, Zlobec I, Pallante P, Sepe R, Esposito F, Zimmermann A, Diamantis I, Terracciano L, Fusco A, Karamitopoulou E. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology 2012; 60:397-404; PMID:22276603; http://dx.doi.org/ 10.1111/j.1365-2559.2011.04121.x [DOI] [PubMed] [Google Scholar]
- 17.Califano D, Pignata S, Losito NS, Ottaiano A, Greggi S, De Simone V, Cecere S, Aiello C, Esposito F, Fusco A, et al.. High HMGA2 expression and high body mass index negatively affect the prognosis of patients with ovarian cancer. J Cell Physiol 2014; 229:53-9; PMID:23765903; http://dx.doi.org/ 10.1002/jcp.24416 [DOI] [PubMed] [Google Scholar]
- 18.Wu ZY, Wang SM, Chen ZH, Huv SX, Huang K, Huang BJ, Du JL, Huang CM, Peng L, Jian ZX, et al.. MiR-204 regulates HMGA2 expression and inhibits cell proliferation in human thyroid cancer. Cancer Biomark 2015; 15:535-42; PMID:26406941; http://dx.doi.org/ 10.3233/CBM-150492 [DOI] [PubMed] [Google Scholar]
- 19.Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH, Kang H, Kim G, An HJ. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett 2015; 356:937-45; PMID:25444913; http://dx.doi.org/ 10.1016/j.canlet.2014.11.011 [DOI] [PubMed] [Google Scholar]
- 20.Kozaki K, Inazawa J. Tumor-suppressive microRNA silenced by tumor-specific DNA hypermethylation in cancer cells. Cancer Sci 2012; 103:837-45; PMID:22320679; http://dx.doi.org/ 10.1111/j.1349-7006.2012.02236.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada K, Baba Y, Ishimoto T. Suppressor microRNA-145 is epigenetically regulated by promoter hypermethylation in esophageal squamous cell carcinoma. Anticancer Res 2015; 35:4617-24; PMID:26254350 [PubMed] [Google Scholar]
- 22.King CE, Wang L, Winograd R, Madison BB, Mongroo PS, Johnstone CN, Rustgi AK. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 2011; 30:4185-93; PMID:21625210; http://dx.doi.org/ 10.1038/onc.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Li XJ, Qiao L, Shi F, Liu W, Li Y, Dang YP, Gu WJ, Wang XG, Liu W. miR-98 suppresses melanoma metastasis through a negative feedback loop with its target gene IL-6. Exp Mol Med 2014; 46:e116; PMID:25277211; http://dx.doi.org/ 10.1038/emm.2014.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Sun Z, Zhu Z, Zhang J, Sun X, Xu H. PBX3 is overexpressed in gastric cancer and regulates cell proliferation. Tumour Biol 2014; 35:4363-8; PMID:24375258; http://dx.doi.org/ 10.1007/s13277-013-1573-6 [DOI] [PubMed] [Google Scholar]
- 25.Ramberg H, Grytli HH, Nygård S, Wang W, Ögren O, Zhao S, Løvf M, Katz B, Skotheim RI, Bjartell A, et al.. PBX3 is a putative biomarker of aggressive prostate cancer. Int J Cancer 2016; 139(8):1810-20. [Epub ahead of print]; PMID:27273830; http://dx.doi.org/ 10.1002/ijc.30220 [DOI] [PubMed] [Google Scholar]
- 26.Han HB, Gu J, Ji DB, Li ZW, Zhang Y, Zhao W, Wang LM, Zhang ZQ. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J Gastroenterol 2014; 20:18260-70; PMID:25561793; http://dx.doi.org/ 10.3748/wjg.v20.i48.18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauhan D, Li G, Hideshima T, Podar K, Shringarpure R, Mitsiades C, Munshi N, Yew PR, Anderson KC. Blockade of ubiquitin-conjugating enzyme CDC34 enhances anti-myeloma activity of Bortezomib/Proteasome inhibitor PS-341. Oncogene 2004; 23:3597-602; PMID:15094775http://dx.doi.org/ 10.1038/sj.onc.1207458 [DOI] [PubMed] [Google Scholar]
- 28.Wong SC, He CW, Chan CM, Chan AK, Wong HT, Cheung MT, Luk LL, Au TC, Chiu MK, Ma BB, et al.. Clinical significance of frizzled homolog 3 protein in colorectal cancer patients. PLoS One 2013; 8:e79481; PMID:24255701; http://dx.doi.org/ 10.1371/journal.pone.0079481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, et al.. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem 2013; 379:69-75; PMID:23591597; http://dx.doi.org/ 10.1007/s11010-013-1628-4 [DOI] [PubMed] [Google Scholar]
- 30.Sterenczak KA, Eckardt A, Kampmann A, Willenbrock S, Eberle N, Länger F, Kleinschmidt S, Hewicker-Trautwein M, Kreipe H, Nolte I, et al.. HMGA1 and HMGA2 expression and comparative analyses of HMGA2, Lin28 and let-7 miRNAs in oral squamous cell carcinoma. BMC Cancer 2014; 14:694; PMID:25245141; http://dx.doi.org/ 10.1186/1471-2407-14-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Ha S, Jung CK, Lee HH. High-mobility-group A2 overexpression provokes a poor prognosis of gastric cancer through the epithelial-mesenchymal transition. Int J Oncol 2015; 46:2431-8; PMID:25845850; http://dx.doi.org/ 10.3892/ijo.2015.2947 [DOI] [PubMed] [Google Scholar]
- 32.Xia YY, Yin L, Tian H, Guo WJ, Jiang N, Jiang XS, Wu J, Chen M, Wu JZ, He X. HMGA2 is associated with epithelial-mesenchymal transition and can predict poor prognosis in nasopharyngeal carcinoma. Onco Targets Ther 2015; 8:169-76; PMID:25653540; http://dx.doi.org/ 10.2147/OTT.S74397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res 2013; 73:4289-99; PMID:23722545; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 2005; 9:403-14; PMID:16139228; http://dx.doi.org/ 10.1016/j.devcel.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzur G, Israel A, Levy A, Benjamin H, Meiri E, Shufaro Y, Meir K, Khvalevsky E, Spector Y, Rojansky N, et al.. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS One 2009; 4:e7511; PMID:19841744; http://dx.doi.org/ 10.1371/journal.pone.0007511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papaioannou G, Inloes JB, Nakamura Y, Paltrinieri E, Kobayashi T. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc Natl Acad Sci U S A 2013; 110:E3291-300; PMID:23940373; http://dx.doi.org/ 10.1073/pnas.1302797110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 2008; 7:3935-42; PMID:19098426; http://dx.doi.org/ 10.4161/cc.7.24.7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowe RG, Wang LD, Coma S, Han A, Mathieu R, Pearson DS, Ross S, Sousa P, Nguyen PT, Rodriguez A, et al.. Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis. J Exp Med 2016; 213(8):1497-512; PMID:27401346; http://dx.doi.org/ 10.1084/jem.20151912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metlapally R, Gonzalez P, Hawthorne FA, Tran-Viet KN, Wildsoet CF, Young TL. Scleral micro-RNA signatures in adult and fetal eyes. PLoS One 2013; 8(10):e78984; PMID:24205357; http://dx.doi.org/ 10.1371/journal.pone.0078984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


