Abstract
Background
The Resistance-Nodulation-Division (RND) family transporter AcrB plays a major role in the intrinsic and increased resistance of Escherichia coli to a large number of antibiotics. The distal binding pocket within this multidrug efflux transporter is very large, but the effort to define the roles of various residues facing this pocket through site-directed mutagenesis so far involved only the determination of minimal inhibitory concentrations of drugs in mutants.
Methods
We measured in intact E. coli cells the kinetics of efflux of two substrates, nitrocefin (a cephalosporin) that is predicted mainly to bind to the upper, “groove” domain of the pocket, and L-alanyl-β-naphthylamide (Ala-Naph) that is likely to bind to the lower, “cave” domain, in a number of site-directed mutants of AcrB, where a hydrophobic or aromatic residue was changed into alanine.
Results
The efflux of nitrocefin became attenuated by some mutations in the groove domain, such as I278A and F178A, but in some experiments a mutation in the cave domain, F628A produced a similar result. In some cases an increased value of KM was detected. The efflux of Ala-Naph was increased by mutations in the cave domain, such as F136A and I626A, but also by those in the groove domain (I277A, I278A, F178A). In most cases the increased Vmax values appeared to be responsible. F610A mutation had a profound effect on the efflux of both substrates, as reported earlier.
Conclusions
Our data show for the first time effects of various substrate-binding pocket mutations on the kinetics of efflux of two substrates by the AcrB pump. They also confirm interactions between substrates and drugs predicted by MD simulation studies, and also reveal areas that need future research.
Keywords: multidrug efflux, RND pump, efflux kinetics, substrate-binding site, site-directed mutagenesis
The global emergence of multidrug-resistant Gram-negative bacteria is currently a major threat to human health [1]. Multidrug efflux pumps, exemplified by the AcrB-AcrA-TolC complex of Escherichia coli, not only play a major role in this multidrug resistance but also act synergistically with other mechanisms of resistance [2]. Although the binding of substrate drugs and inhibitors to the RND (Resistance-Nodulation-Division) transporter AcrB has been studied by crystallography [3-6], molecular docking [7], as well as molecular dynamics simulations [8], an early effort to use site-directed mutagenesis found that only F610A mutation, in the distal (or deep) binding pocket of AcrB, produced a change in the efflux of many drugs [9]. A ciprofloxacin-resistant clinical isolate of Salmonella Typhimurium was found to contain a G288D mutation in the upper part of the distal binding pocket of AcrB, which apparently improves the efficiency of ciprofloxacin efflux but adversely affects the efflux of minocycline and doxorubicin [10]; however an identical change in Escherichia coli AcrB was not found to alter the drug efflux phenotype [11].
In most of these studies, drug efflux was inferred from alterations in minimal inhibitory concentrations (MICs) of the drugs. This approach, however, is very crude, because MICs are the results of interactions of many processes, including influx, efflux, drug inactivation, and drug binding, and they are unlikely to be the direct indicator of the efflux process alone. We have, however, succeeded in determining the kinetics of the efflux of two types of substrates, β-lactams [12, 13] and aminoacyl-β-naphthylamides [14]. We have generated AcrB mutants in which various residues in the substrate-binding cavities were converted to alanine, and we have directly determined the effect of these mutations on efflux kinetics, by using representatives of these two types of substrates, nitrocefin (a cephalosporin) and Ala-β-naphthylamide or Ala-Naph (an aminoacyl-β-naphthylamide). The results shed light on the way substrates become bound in the extremely large cavity of the distal binding pocket of AcrB, and then become extruded by the conformational change(s) produced by the translocation of proton through its transmembrane domain.
EXPERIMENTAL PROCEDURES
Selection of Residues to be Mutated
Most of the residues converted to Ala are those facing the distal binding pocket as defined previously [7]. Additionally Q176 and I278 were mutated because these residues appeared to interact with nitrocefin in the molecular dynamics simulations [8].
Efflux Assays
For the determination of nitrocefin efflux, strain HN1159 (RAM121 ΔacrR::kan ΔacrAB::spc)[13] containing pACYC184acrAB (with desired mutations in acrB) was used. Details on this plasmid can be found in our previous publication [11]. Mutations were introduced in the acrB gene by site-directed mutagenesis using a QuickChange Lightning Site Directed Mutagenesis kit (Agilent Technologies). RAM121 is a strain with a mutated OmpC porin, which produces an abnormally large channel [15]. This large channel accelerates the influx of large, hydrophobic substrates such as nitrocefin (or Ala-Naph), facilitating the quantitation of their efflux. In this setting, the hydrolysis of nitrocefin was catalyzed by the chromosomal AmpC β-lactamase of E. coli.
For the determination of Ala-Naph efflux, ΔpepN::kan mutation was first transduced into RAM121 from JW0915-1 in Keio collection (obtained from E. coli genetic stock center, Yale University), followed by the transduction of ΔacrAB::spc [14]. RAM121 ΔpepN::kan ΔacrAB::spc was then transformed with pMAL-PepN [14] and the transformants were selected on 100 μg/ml ampicillin plates at 37°C. This transformant was then transformed with pACYC184 acrAB (with or without a mutation within AcrB) followed by selection on LB plates containing both ampicillin (100 μg/ml) and chloramphenicol (35 μg/ml). RAM121 ΔpepN::kan ΔacrAB::spc containing both pMAL-PepN and pACYC184 acrAB was then grown at 30°C in modified LB broth (1% tryptone, 0.5% yeast extract, 0.87% NaCl and 5 mM MgSO4) containing 100 μg/ml ampicillin, 35 μg/ml chloramphenicol and 0.1% glucose until the culture reached an OD600 of 0.5. Cells were harvested by centrifugation, washed twice and resuspended in 50 mM potassium phosphate buffer (pH 7) containing 5 mM MgCl2 to an OD600 of 0.8. The cell suspension was then diluted (1:8) in the same buffer. The Ala-Naph efflux assay was carried out as described previously [14]. The external concentrations of Ala-Naph varied usually up to 1.0 mM.
Results of efflux assays were analyzed by non-linear regression using program CurveExpert (www.curveexpert.net).
RESULTS
Classification of Residues
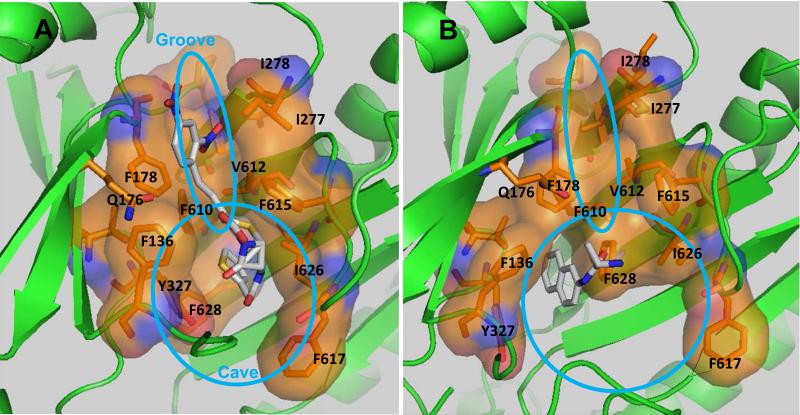
Earlier docking studies from our laboratory [7] showed that a majority of substrates bound to the narrow “groove” region (at the top of picture in Fig. 1), corresponding to the portion closer to the exit gate of the distal binding pocket of AcrB, whereas some other substrates, such as ethidium and chloramphenicol, bound to the wider “cave” region at the bottom of the pocket (also shown in Fig. 1). We have selected from the groove region residues Q176, I277, I278, and V612, in part on the basis of their expected interaction with nitrocefin in molecular dynamics simulations [8]. The residue F178 is shared by both the groove and cave regions, being situated at the entrance of these subdomains. The cave region, lined with many Phe residues (F136, F628, F610, F615, F617) and other hydrophobic residues including I626 and Y327, corresponds to what was recently called “hydrophobic trap” in the study of the binding of inhibitors to AcrB and its relatives [4]. Molecular dynamics simulations [8] gave results largely consistent with these classifications. In addition, the efflux of nitrocefin, a typical groove binder, was inhibited by the simultaneous presence of minocycline, which is known to bind to the groove area from the co-crystal study [3], docking [7], and MD simulations [8], whereas it was not inhibited at all by the simultaneous presence of chloramphenicol [7]. Chloramphenicol behaved as a cave binder in docking [7], although in MD simulations it tended to come out from the cave and enter at least partially into the groove area [8]. These different binding behaviors of substrates are consistent with the idea that substrates tend to bind to one of the two sub-areas of the distal binding pocket. More recently, some of the aminoacyl-β-naphthylamides, including Ala-Naph, was found to bind mostly to the hydrophobic pocket or the cave [14]. We therefore tested quantitatively the effect of mutations in the distal binding pocket on the efflux of both a groove binder, nitrocefin, and a cave binder, Ala-Naph.
Fig. 1.
The distal binding pocket in the binding protomer of AcrB. A and B represent the near-equilibrium structures obtained after lengthy molecular dynamics simulations of nitrocefin and Ala-Naph bound to the wild-type AcrB [8, 14]. The areas defined as groove and cave are loosely indicated as ellipses.
The alteration of one residue, F610A, had a strong effect on the efflux of most substrates, as was shown by the MIC data by Bohnert and others [9] and by the nitrocefin efflux assay [11]. This residue appears to play a special role in the efflux process (see Discussion).
Efflux of Nitrocefin
Nitrocefin is pumped out by the wild type AcrB with a Michaelis-Menten kinetics, with the Vmax of about 0.02 nmol/mg/s and the KM of around 5 μM [13], which were confirmed here also (Table 1, Fig. 2). The Vmax in our present system was slightly higher than previously reported (0.039 rather than 0.023 nmol/mg/s), presumably due to the fact that AcrB was expressed from plasmids rather than from chromosome.
Table 1.
Kinetic Constants of Ala-Naph and Nitrocefin Efflux by AcrB
| AcrB | Nitrocefin | Ala-Naph | |||||
|---|---|---|---|---|---|---|---|
| Cp (μM) at 175 μM Co | KM (μM) | Vmax (nmol/s/mg) | Cp at 1 mM Co | K0.5 (μM) | Vmax (nmol/s/mg) | Hill coeff. | |
| Wild Type | 19.5 (6)a | 3.8±1.0 | 0.039±0.006 | 268 (4) | 176±19 | 82±16 | 1.7±0.1 |
| Q176A | 21.4 (2) | 1.1,2.6 | 0.033,0.039 | 263 (3) | 177±45 | 105±44 | 1.7±0.2 |
| I277A | 20.2 (6) | 3.9±2.1 | 0.034±0.004 | 144 (4) | 143±27 | 147±104 | 2.0±0.6 |
| I278A | 30.9 (5) | 2.7±1.3 | 0.025±0.001 | 77 (5) | >300 | >200 | |
| V612A | 22.7 (3) | 4.3±1.3 | 0.033±0.001 | b | 172±46 (5) | 93±39 | 2.0±0.2 |
| F178A | 32.9 (3) | 14.5±1.2 | 0.04±0.02 | 62 (3) | >300 | >200 | |
| F136A | 24.5 (4) | 3.1±1.6 | 0.03±0.003 | 114 (2) | 138,172 | 194,289 | 1.6,1.5 |
| F610A | 38.5 (5) | 1,030±400c | 3.1±1.2 | 154 (5) | 92±16 | 110±41 | 2.7±0.2 |
| F615A | 24.1 (4) | 4.3±1.9 | 0.03±0.002 | b | 106±50 (3) | 217±148 | 1.8±0.7 |
| I626A | 20.3 (4) | 2.5±0.9 | 0.033±0.003 | 134 (4) | 255±77 | 268±44 | 1.5±0.1 |
| F617A | 9.5 (6) | 2.9±1.0 | 0.052±0.005 | 213 (4) | 135±21 | 102±34 | 2.0±0.4 |
| F628A | 22.0 (3) | 5.5±2.9 | 0.036±0.007 | 287 (2) | 145,143 | 73,74 | 1.9,2.0 |
Values shown are average with the number of experiments in parenthesis. Values that are significantly different from that in the wild type are shown in bold. For kinetic constants, standard deviations are also shown, when possible.
Not available as the maximum Co used was 0.5 mM.
Kinetics became sigmoidal in this mutant, and thus this value represents K0.5 rather than KM.
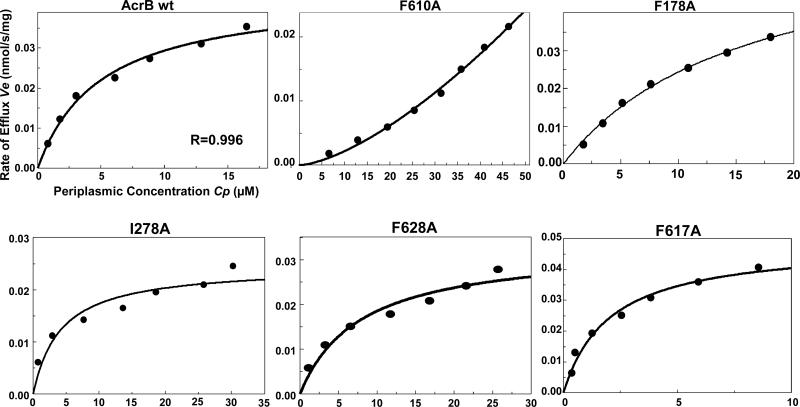
Fig. 2.
Some representative data for nitrocefin efflux assay. Since only small changes were seen with most mutants, only those showing large changes are shown together with the data for the wild type. With F628A, the effect was not always reproducible (see text).
In assessing the effect of various mutations on nitrocefin efflux, one useful criterion is its periplasmic concentration Cp at a fixed external concentration Co (Table 1), because if the pump becomes less efficient Cp will become higher than in the presence of the wild type AcrB. This criterion is also advantageous in that we do not suffer from the uncertainties involved in non-linear regression analysis. Clearly, the efflux is less efficient in three mutants, I278A, F178A, and F610A, and is surprisingly more efficient in F617A (Table 1, Fig. 2). In the case of F178A, the KM value is strongly increased (Table 1). The lowered activity of I278A and F178A mutants is satisfying as these are the residues that interact with the dinitrophenyl group of nitrocefin (Fig. 1A) in the upper “groove” subdomain, where most of the nitrocefin molecule binds. We have earlier reported [11] that F610A mutation produces a drastic change in the nitrocefin efflux kinetics, which now becomes clearly sigmoidal with a very large K0.5 value (around 1 mM) and a large estimated Vmax (around 3 nmol/mg/s), with a Hill coefficient of about 1.5. Among other mutants in the cave region, F628A was less effective in nitrocefin efflux in 2 out of 3 assays (Fig. 2), although this is not seen in the average Cp value in Table 1 because of the data in the other assay. This effect of F628A mutation was somewhat unexpected from the MD simulation generated model (Fig. 1A), as F628 was not very close (about 6 Å) to the nitrocefin thiophene ring. We speculate on the behavior of F617A in Discussion.
Efflux of Ala-Naph
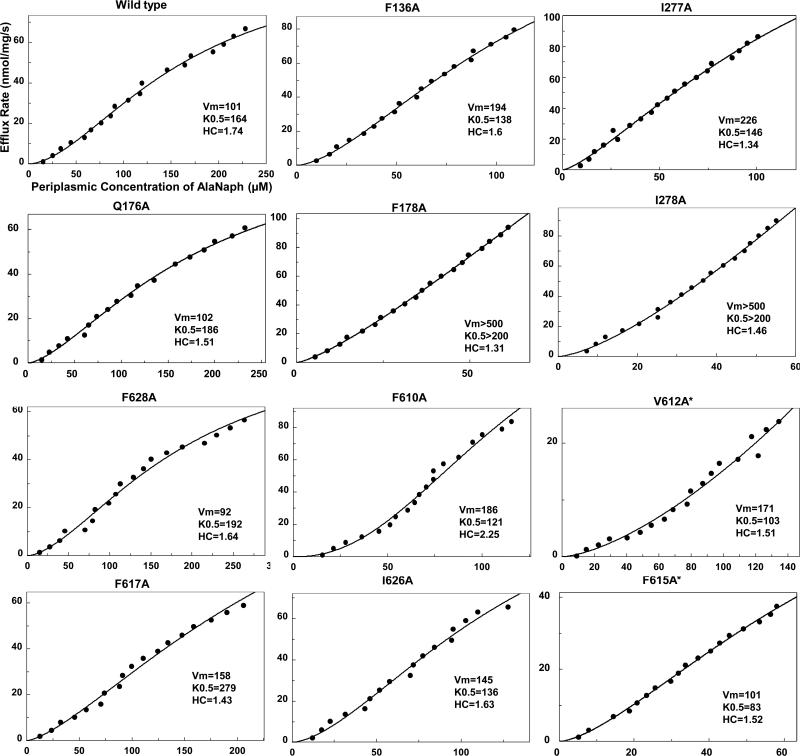
Ala-Naph is pumped out very rapidly (Vmax of 82 nmol/mg/s) by the wild type AcrB with sigmoidal kinetics with the K0.5 of 176 μM and the Hill coefficient of 1.7 (Table 1). When we examined Ala-Naph efflux in mutants, some of them were surprisingly found to produce increased efflux, rather than the expected decreases. The increases in Ala-Naph efflux are again seen most convincingly in the decreased periplasmic concentration of Ala-Naph (Cp) at the given external concentration (Co) of the substrate, in comparison with the wild-type AcrB (Table 1). Most of the mutations that produced a more effective Ala-Naph efflux were located in the cave subdomain, a satisfying outcome because Ala-Naph is bound to the cave in MD simulations (Fig. 1). These mutations include F136A, F610A, I626A, and likely F615A, and F178A shared by the cave and the groove (Table 1, Fig. 3). In most cases the decreased values of K0.5 and/or increased values of Vmax appeared to be responsible for this change of efflux kinetics (Table 1). In F610A, the Hill coefficient is also clearly increased. However, mutations that altered Ala-Naph efflux were not completely limited to the cave subdomain, and a few alterations in the groove subdomain (I277A, I278A) also led to the accelerated Ala-Naph efflux (Table 1, Fig. 3). The changes in I278A were rather extreme, and we have not been able to get convincing estimates of kinetic constants as both Vmax and K0.5 appeared to have very large values (Fig. 3).
Fig. 3.
Representative data for Ala-Naph efflux assay. Note that the values of kinetic constants shown in Table 1 are averages of those obtained in individual experiments, and thus are different from those shown here.
Discussion
We have examined the detailed kinetics of drug efflux by mutants of a prototypical multidrug transporter AcrB of E. coli. The substrates used were nitrocefin, which binds mainly to the upper, “groove” domain of the very large distal binding pocket [16], and Ala-Naph, which binds mostly to the lower, “cave” domain of the pocket [14] (Fig. 1). We have found earlier, by both docking [7] and molecular dynamics simulations [8], that many common antibiotics, including nitrocefin and minocycline, bind predominantly to the groove area, which contains numerous hydrophilic and charged residues. (It should be noted that with large, elongated substrates such as nitrocefin, its hydrophobic end containing the thiophene ring actually binds to the cave area [see Fig. 1A]). We also know that the efflux of nitrocefin is inhibited by the simultaneous presence of minocycline, but not of chloramphenicol [7], whose binding preference is either the cave or the cave/groove interface [7,8]. We found in this study that the efflux of nitrocefin was not visibly altered by most mutations in the cave area or even the groove area. Perhaps this is expected as these areas contain many residues that could furnish alternative interactions with this compound that contains a negative charge, several atoms capable of H-bonding, and locally high dipolar moments in the dinitrophenyl group. One exception was the F610A mutation that had a profound effect of changing the Michaelis-Menten kinetics to a sigmoidal one, as reported earlier [11]. Molecular dynamics simulation suggested that the main effect of this mutation is to affect the conformational transition of AcrB needed for drug export [17], and this emphasizes the possible difficulty in interpreting mutant data. A mutation that raised the value of KM significantly was F178A, the residue that straddles the groove and cave areas, and I278A in the groove also made nitrocefin efflux less efficient, apparently by reducing the Vmax (Table 1, Fig. 2). A mutation in the cave area, F617A, surprisingly made nitrocefin efflux more efficient. F617 is very far away from the bound nitrocefin molecule (Fig. 1), but is at the tip of the glycine-rich “switch loop” thought to control the entry of ligands to the distal binding pocket [5]. Thus its conversion to a much smaller Ala residue may speed up the entry of nitrocefin with its elongated shape into the pocket.
In contrast, the efflux of Ala-Naph, for which molecular dynamics simulation predicts binding of the naphthalene ring to the strongly hydrophobic cave area [14] (Fig. 1B), was significantly affected by various mutations, mostly in the cave area (Table 1), confirming the importance of the cave as the substrate binding area, especially for lipophilic ligands such as solvents [7, 16] and acyl naphthylamides with small substituents, such as Ala-Naph [14]. However, surprisingly these mutations enhanced, rather than diminished, Ala-Naph efflux, perhaps suggesting that a too tight binding to the cave could actually slow down the efflux. Indeed the binding energy calculated for Ala-Naph was −22.9 kcal/mol [14], much larger in size than the binding energy calculated for solvents, such as benzene (−11.0 kcal/mol [16]), that are thought to be pumped out at a very high rate [16].
An earlier study [4] proposed that this area should be named “hydrophobic trap” “that branches off the substrate translocation channel,” and suggested that the binding to this area is limited to inhibitors. However, considering this area as a pure trap may be an exaggeration. Indeed the entire cave area (in addition to the groove subdomain) becomes collapsed in the Extrusion protomer of AcrB [3], and this suggests that the cave may function as a legitimate substrate-binding pocket in efflux, consistent with our results on Ala-Naph. Nevertheless, our observation that less tight binding to this cave area accelerates Ala-Naph efflux is not inconsistent with the idea of a “trap.” A very tight binding (seen with some inhibitors) to this area may thus essentially stop the export, explaining the behavior of some inhibitors that at least partially bind to this domain [4, 6, 18], and also the acceleration of efflux of Ala-Naph in some of the cave mutants in which the binding may have become less tight (Table 1, Fig. 3).
This interpretation is also consistent with previous computational studies on the effect of the F610A substitution on the binding to the groove of doxorubicin and minocycline [17, 19], which showed that the removal of the bulky phenyl ring induced sliding of both compounds within the Phe-rich cave, increasing their binding affinity and thus slowing down efflux. While both doxorubicin and minocycline are groove binders sitting just above F610, nitrocefin with its elongated shape is in contact with the cave area (see above), and Ala-Naph is mainly bound to that area. Therefore it is not surprising that the F610A substitution has a different effect on these substrates; for example, it is reasonable to expect that the substitution will not induce a large displacement of nitrocefin and Ala-Naph, as seen for doxorubicin and minocycline.
Highlights.
RND efflux pumps such as AcrB make a major contribution to multidrug resistance in gram-negative bacteria.
Drugs are bound to either the groove or the cave subdomains of the large distal binding pocket of AcrB.
Site-directed mutagenesis should reveal the details of interaction between the drugs and the pocket.
We have determined the efflux kinetics of nitrocefin (a groove binder) and L-alanyl-β-naphthylamide (a cave binder) in mutants.
Mutations often attenuated nitrocefin efflux yet sometimes accelerated the naphthylamide efflux.
Acknowledgments
FUNDING. This study was supported by the National Institutes of Health (grant 5 R01 AI-009644) in Berkeley, and by Translocation Consortium in Italy. The research of A.V.V. leading to these results was conducted as part of the Translocation consortium (www.translocation.com) and received support from the Innovative Medicines Initiatives Joint Undertaking under Grant 115525, resources of which are composed of a financial contribution from the European Union's seventh framework program (FP7/ 2007–2013) and European Federation of Pharmaceutical Industries and Association companies’ kind contribution.
Abbreviations
- RND
resistance-nodulation-division
- Ala-Naph
L-alanyl-β-naphthylamide
- MD
molecular dynamics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Centers for Disease Control and Prevention . Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention; Atlanta, GA: 2013. http://www.cdc.gov/drugresistance/threat-report-2013/index.html. [Google Scholar]
- 2.Li X-Z, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Revs. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 4.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 5.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim H-S, Ding X, Ornik AR, Ruggerone P, Bowlin TL, Nikaido H, Pos KM, Opperman TJ. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohnert JA, Schuster S, Seeger MA, Fahnrich E, Pos KM, Kern WV. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blair JM, Bavro VN, Ricci V, Modi N, Cacciotto P, Kleinekathfer U, Ruggerone P, Vargiu AV, Baylay AJ, Smith HE, Brandon Y, Galloway D, Piddock LJ. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3511–3516. doi: 10.1073/pnas.1419939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soparkar K, Kinana AD, Weeks JW, Morrison KD, Nikaido H, Misra R. Reversal of the Drug Binding Pocket Defects of the AcrB Multidrug Efflux Pump Protein of Escherichia coli. J. Bacteriol. 2015;197:3255–3264. doi: 10.1128/JB.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim SP, Nikaido H. Antimicrob. Vol. 54. Agents Chemother; 2010. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli; pp. 1800–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinana AD, Vargiu AV, May T, Nikaido H. Aminoacyl beta-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1405–1410. doi: 10.1073/pnas.1525143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra R, Benson SA. Isolation and characterization of OmpC porin mutants with altered pore properties. J. Bacteriol. 1988;170:528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinana AD, Vargiu AV, Nikaido H. Some ligands enhance the efflux of other ligands by the Escherichia coli multidrug pump AcrB. Biochemistry. 2013;52:8342–8351. doi: 10.1021/bi401303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargiu AV, Collu F, Schulz R, Pos KM, Zacharias M, Kleinekathofer U, Ruggerone P. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J. Am. Chem. Soc. 2011;133:10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 18.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014;58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggerone P, Murakami S, Pos KM, Vargiu AV. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]