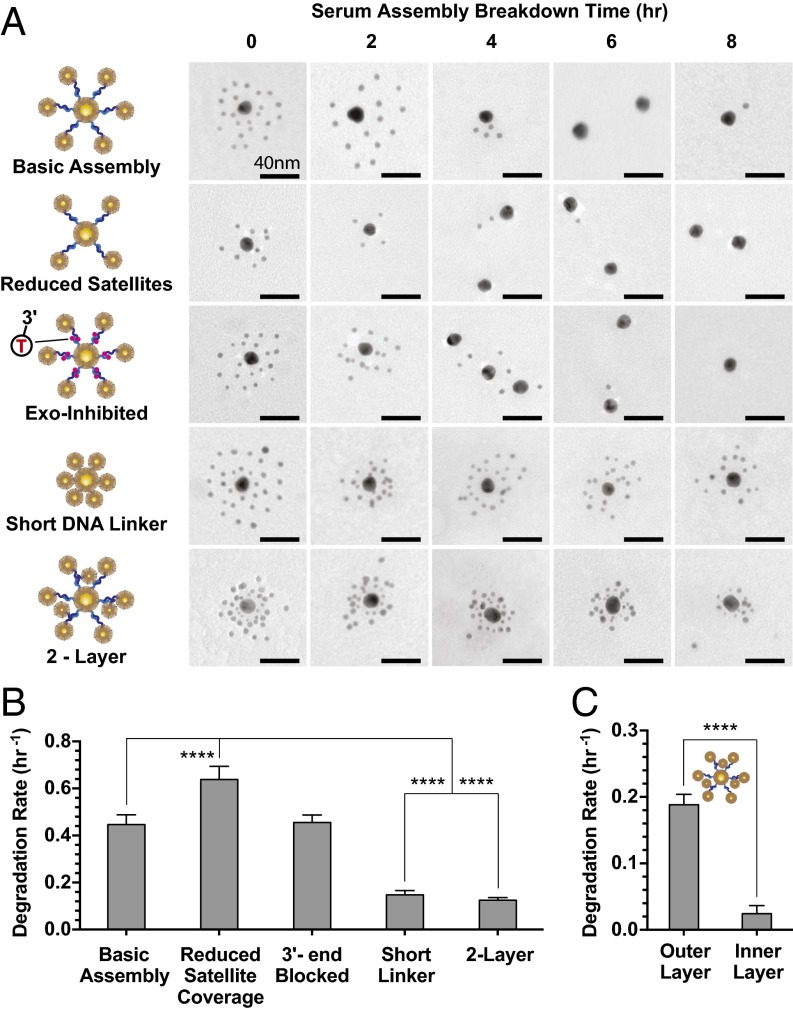
Fig. 4.
Controlling serum degradation rates of GNP-DNA assemblies. (A) Representative TEM (zoomed out TEM is shown in SI Appendix, Figs. S22–S26) of five assembled structures (details are in SI Appendix, Fig. S18) incubated in serum. Two-layer structure: 5 kDa PEG; all other structures: 10 kDa PEG. (B) Associated serum degradation rates. Number of satellites per core nanoparticle quantified by direct TEM counting. Degradation rates are based on single-phase decay fits of these data (SI Appendix, Fig. S19A). Increase in satellite density correlates with improved protections against serum breakdown. (C) Degradation rates of the two-layer structures; 5-nm satellites in the outer layer were quantified separately from the inner-layer 3-nm satellites. Degradation of the outer layer occurs much more rapidly that of the inner layer. Degradation rates are based on single-phase decay fits of these data (SI Appendix, Fig. S19B); error bars are 95% fitting confidence intervals. Statistical significance was determined by (B) ordinary one-way ANOVA or (C) unpaired t test. ****P ≤ 0.0001.