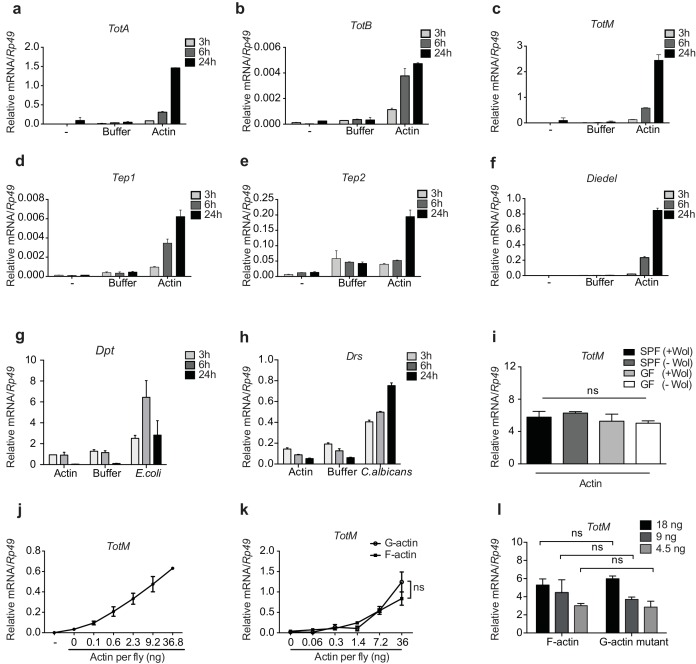
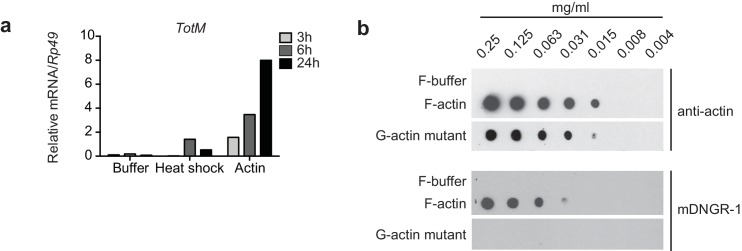
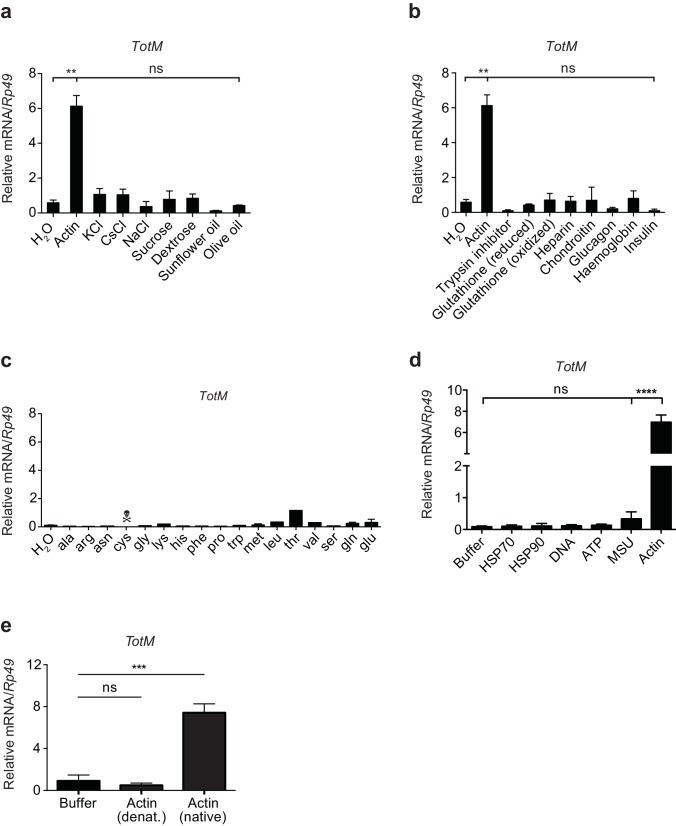
Figure 2. G- and F-actin induce a unique sterile inflammatory response upon injection into Drosophila.
Control (w1118) flies were mock treated (-) or injected with actin buffer, actin, E. coli or C. albicans, as indicated. Flies were euthanised at 3, 6 and 24 hr post injection, RNA was isolated and relative gene expression determined by quantitative RT-PCR. Depicted are expression levels of (a–c) Turandot (Tot) A, TotB and TotM, (d–e) Thioester-containing protein (Tep) 1 and Tep2, (f) Diedel, (g–h) Diptericin (Dpt) and Drosomycin (Drs). (i) Flies colonised with or free of Wolbachia (Wol) were reared under standard (SPF) or germ-free (GF) conditions. Relative TotM expression was assessed 24 hr post actin injection. (j) Dose-dependent TotM response to injected actin at 24 hr post injection. (k) Dose-response curve for latrunculin B-stabilised G-actin vs phalloidin-stabilised F-actin diluted in G-actin or F-actin buffer, respectively. Relative expression levels of TotM 24 hr post injection are shown. (l) A non-polymerisable G-actin mutant and F-actin were serially diluted in F-buffer at the indicated concentrations before injection into flies. Relative expression levels of TotM 24 hr after injection are depicted. Relative gene expression levels were calculated using the housekeeping gene Rp49 as a reference gene. Data are representative of at least two independent experiments with 10 flies/sample with duplicate samples. Bars represent mean ± SEM. Statistical analysis was performed using two-way ANOVA with Sidak’s multiple comparison test as post-test for pairwise comparisons. Results of Sidak’s multiple comparison test are shown (ns, not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). If not otherwise indicated, flies were injected with 36.8 ng of actin per fly. All data points are plotted even where no bars are visible.