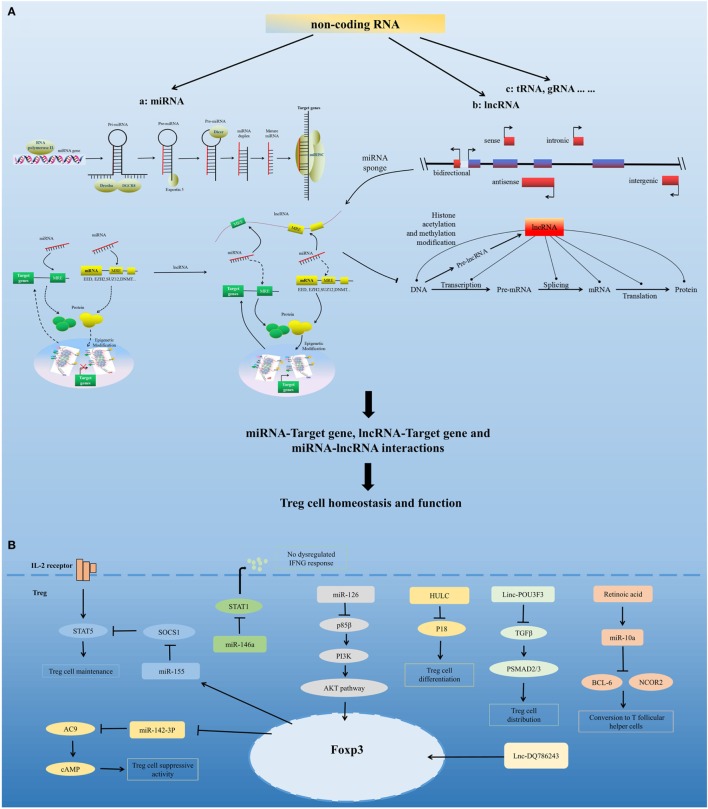
Figure 2.
Non-coding RNA-mediated regulation of Treg cell homeostasis and function. (A) Model for non-coding RNAs (ncRNA)-mediated regulation of Treg cell homeostasis and function. ncRNAs include highly abundant and functionally important RNAs, such as microRNAs (miRNA), long non-coding RNAs (lncRNA), and tRNA. miRNAs are sequentially processed from longer transcripts by the RNase III enzymes Drosha and Dicer. Pri-miRNAs are processed by Drosha into hairpin structures (pre-miRNAs). Exportin 5 shuttles pre-miRNAs from the nucleus into the cytoplasm, where the RNase III Dicer cleaves off the hairpin loop of the pre-miRNA. The duplex segregates, and the mature single-stranded miRNA associates with argonaute proteins and other accessory proteins to form the miRNA-induced silencing complex, which directly mediates the translational repression and the increased degradation of its mRNA targets. miRNA can also indirectly regulate gene expression by repressing the expression of several key enzymes involved in epigenetic modification processes, such as DNA methylation and histone modification. Based on the position of lncRNA relative to the neighboring protein-coding genes in the genome, lncRNAs can be divided into five categories, namely, sense, antisense, bidirectional, intronic, and intergenic. lncRNAs can modulate chromatin modification, mRNA stability, miRNA activity, and the function of proteins by interacting with chromatin, RNA, and protein. lncRNA functions as a miRNA sponge, sequestering miRNAs to regulate the expression level of other transcripts sharing common miRNA response elements. This process leads to fewer miRNA molecules available to bind to target mRNA and, thus, an increase in its protein expression level. miRNA, lncRNA, and mRNA form a well-regulated interacting network and play critical regulatory roles in Treg cell homeostasis and function. (B) Regulation of Treg cells by several representative miRNAs and lncRNAs, including miR-155, miR-146a, miR-126, miR-10a, miR-142-3p, HULC, Linc-POU3F3, and Lnc-DQ786243.