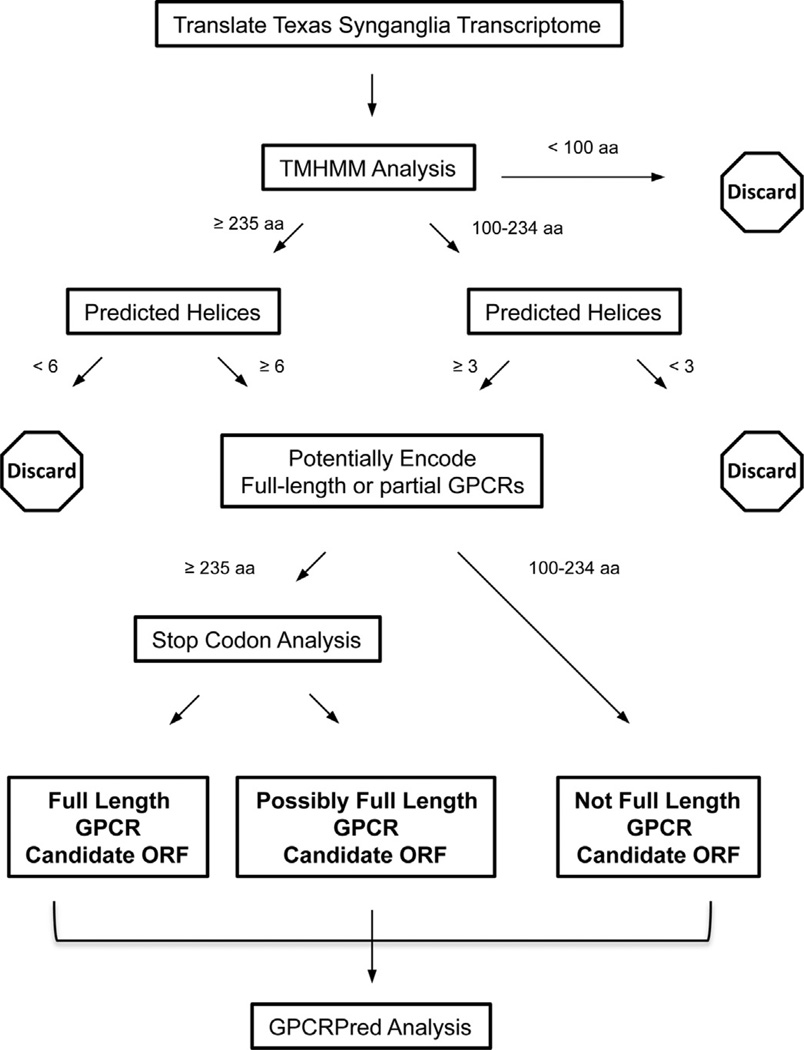
Fig. 2.
Flow chart of ORF prediction process whereby the translated Texas synganglion transcriptome ORFs are routed through TMHMM and GPCRPred to identify candidate GPCR-encoding sequences. The transcriptome sequences were translated into all 6 possible ORFs and those containing <100 amino acids were omitted from further analysis. All remaining ORFs were analyzed by TMHMM to predict and tally transmembrane helix regions. Following TMHMM, ORFs containing <100 aminoacids, ORFs containing 100–234 amino acids and <3 predicted helices, or ORFs containing ≥235 amino acids and <6 helices were omitted from further analysis. The corresponding nucleotide sequences from the remaining ORFs were examined for stop codons in the presumptive 5′ and 3′ untranslated regions to predict if the ORF encodes a full length protein. ORFs containing ≥2 stop codons prior to the putative initiator methionine codon and ≥2 stop codons at the 3′ end of the ORF were parsed as “Full Length GPCR Candidate ORF”. ORFs with only one stop codon both before and after the protein sequence were parsed as “Possibly Full Length GPCR Candidate ORF”. ORFs not meeting these criteria were parsed as “Not Full Length GPCR Candidate ORF”. ORFs in each set were analyzed by GPCRPred.