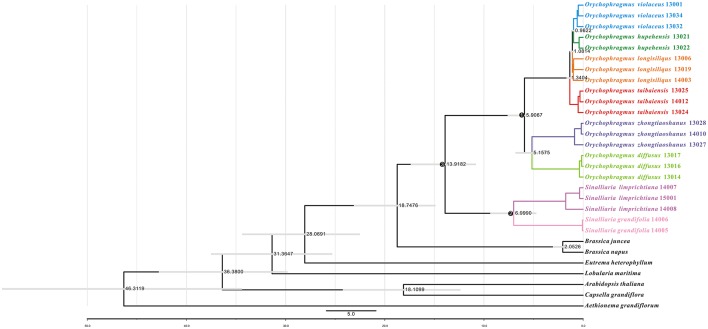
Figure 4.
Divergence time estimates using the average substitution rate based on the whole chloroplast genome sequences. Divergence times of species based on uncorrelated relaxed clock method, using a substitution rate of 5.1952E-4 per base per million years calculated by BEAST program over the whole chloroplast genome sequences. The legend describes the divergence time in million years, and the gray boxes represent the 95% highest probability density of divergence times. In addition, three major nodes were used for divergence comparisons in the Table S3.