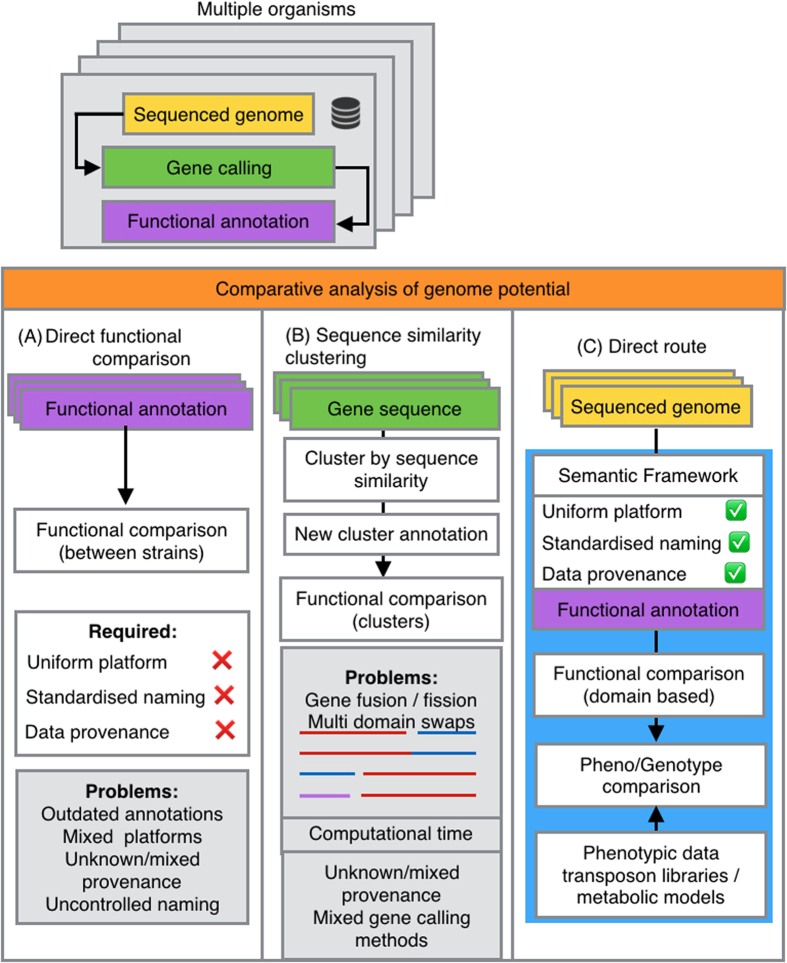
Figure 1. Alternatives for functional genome comparison.
(A) Direct comparison of genome potential using existing annotation is often hampered by lack of standardization of gene calling and annotation tools, mixed and unknown data provenance and inconsistent naming of function. (B) Sequence similarity clustering bypasses inconsistent functional annotations. Computational time scales quadratically with the number of genome sequences and gene fusion/fission events might be overlooked. (C) Usage of standardised annotation tools ensures uniform genome annotation prior to comparison; annotation provenance is stored for all steps.