Figure 3. Domain architectures in sequence based clusters of orthologous proteins.
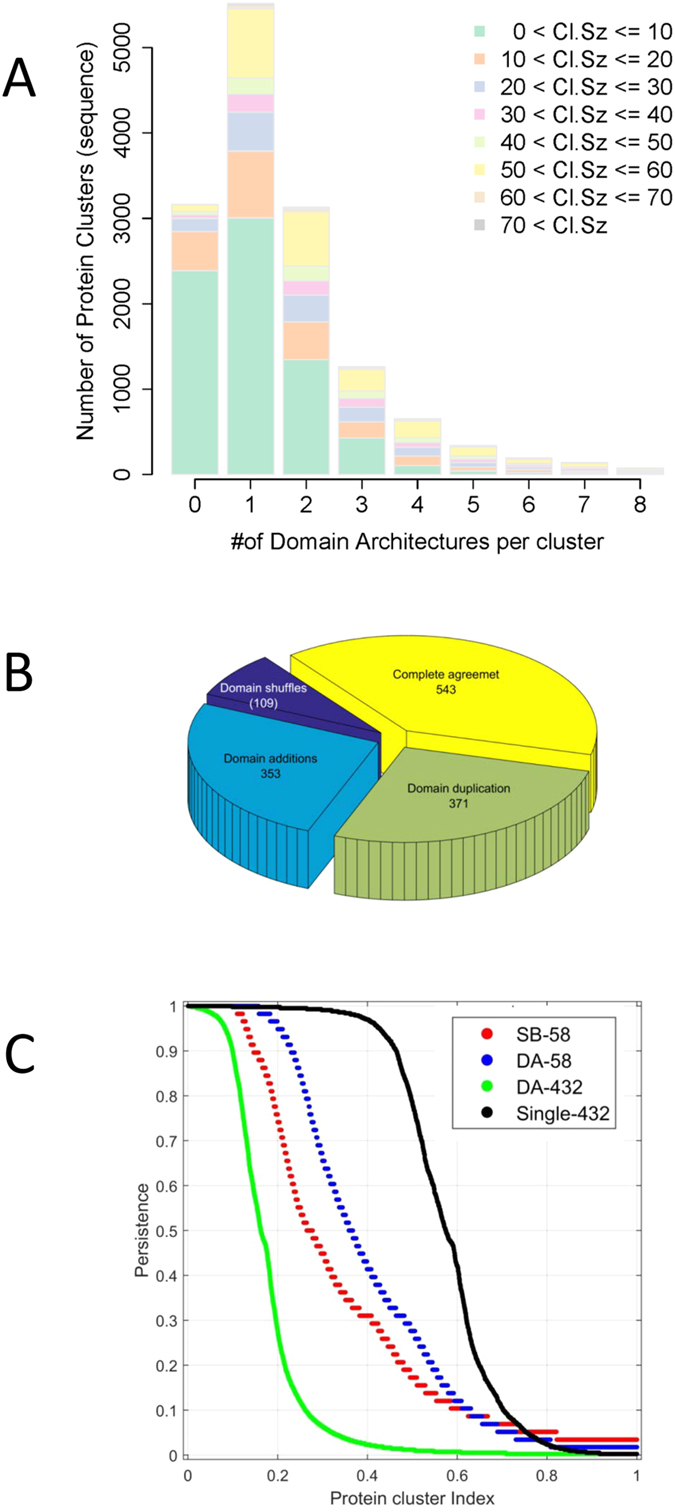
(A) Number of distinct domain architectures per cluster (B) Variability in domain architectures per gene cluster in core-genome. Complete agreement indicates a unique domain architecture shared by all members of the cluster; For the cases where multiple domain architectures were found in a sequence cluster, the number of cases corresponding to domain duplications, additions and shuffles are indicated. (For A and B only 58 complete genome sequences considered). (C) Persistence analysis within the Pseudomonas genus. The curves indicate the persistence of each of the cluster. Clusters have been arranged by decreasing persistence values and the x-axis has been scaled to 0–1 range, in this way the cluster with the highest persistence have an x value of 0 and the cluster with the lowest persistence has an x value of 1. The y-axis indicates the persistence of a given cluster (see Equation 1): for instance a persistence of 0.8 indicates that 80% of the analyzed genomes contain sequences in that given cluster. SB-58 refers to the use of sequence based cluster considering the 58 complete genomes; DA-58 and DA-432 refers to the use of protein domains, for 58 and 432 genomes respectively; Single-432 reproduces the analysis for single domain proteins found in the full set 432 genome sequences.
