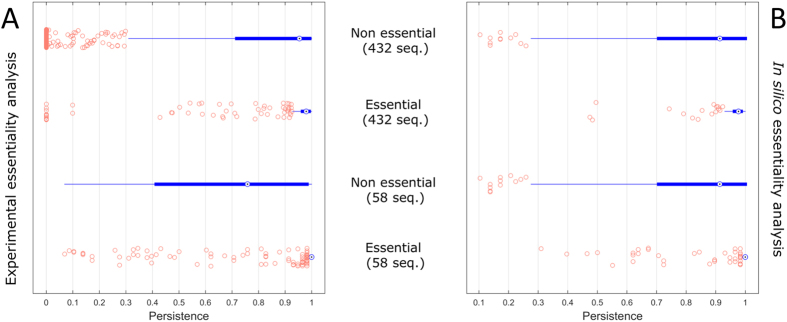
Figure 6. Persistence of (non) essential genes.
(A) Persistence of essential and non-essential genes as derived by experimental investigations. (B) Persistence of essential and non-essential genes as derived by in silico modelling using genome based constrained metabolic modelling. Results shown pertain the use of the iMO1086 model for P. aeruginosa PAO1. In both cases persistence is calculated using the 58 completely sequenced Pseudomonas genomes and the complete set of 432 genomes sequences. Magenta (circle) dots indicate outliers.