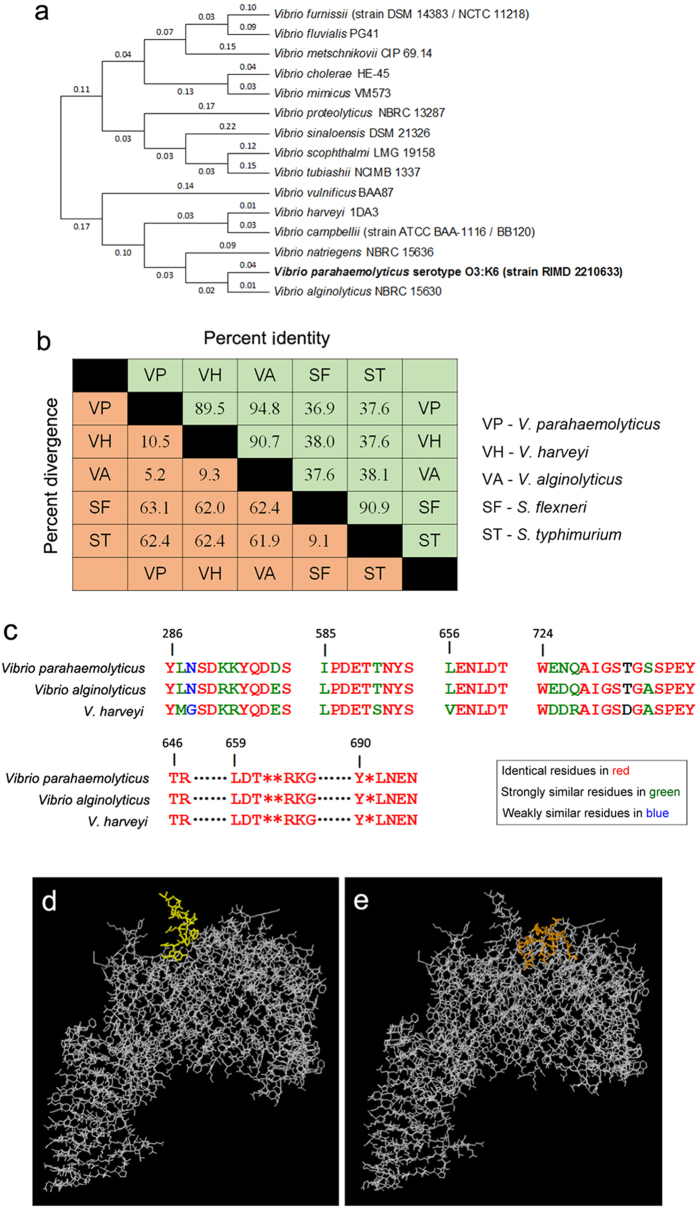
Figure 2. Bioinformatics analysis of LptD based on the amino acid sequences.
(a) A phylogenetic tree based on Lptd sequences was constructed using MEGA software, version 6.0, based on the maximum likelihood method. (b) Percentage similarities and divergences of the LptD sequences from other Gram-negative bacteria. The sequence pair distances of amino acid residues were determined by ClustalW. (c) Homology alignment of four linear antibody epitopes – YLNSDKKYQDDS (286–297) (score of 0.77), IPDETTNYS (585–593) (score of 0.648), LENLDT (656–661) (score of 0.507) and WENQAIGSTGSSPEY (724–738) (score of 0.78) – and the discontinue epitope T646R647…L659D660T661**R664K665G666…Y690*L692N693E694N695 among major Vibrio species. Epitopes of V. parahaemolyticus LptD were predicted by the ElliPro online server, based upon the modelled three-dimensional structure of LptD. Identical residues are in red, strongly similar residues are in green, and weakly similar residues are in blue. The three-dimensional structure shows (d) a representative, predicted linear epitope of WENQAIGSTGSSPEY (724–738) (epitope residues are in yellow) and (e) a discontinuous epitope of T646R647…L659D660T661**R664K665G666…Y690*L692N693E694N695 (epitope residues are in orange).