Figure 1. Secondary and tertiary structural prediction of H. pylori CagL (strain 26695); the selection of ten short motifs for generating deletion mutants is indicated with regard to the recently solved CagL crystal structure.
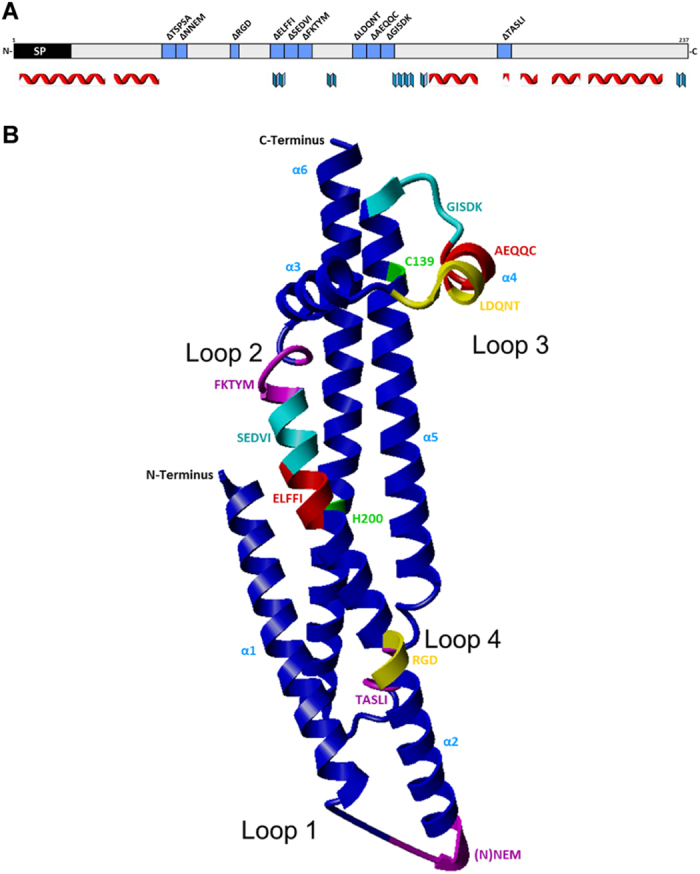
(A) Placement of the ten motif deletion mutants within the secondary structure of CagL. Secondary structure prediction was performed using Jpred (www.compbio.dundee.ac.uk/jpred). Alpha helices are indicated by red symbols, beta sheets are indicated by blue symbols. Only the structured regions of confidence score ≥1 of the Jpred prediction are indicated by symbols below the sequence. SP designates a predicted leader peptide which probably serves to transport CagL to the periplasm. (B) Placement of the ten motif deletion mutants in the CagL crystal structure (pdb: 3zcj_chainA38), which was imaged in ribbon mode by Yasara (www.yasara.org). Motif deletion mutants along the 2D and 3D structure are indicated in blue (2D) and in different colors (3D structure; pink, turqoise, red, purple, yellow) contrasting to the backbone shown in royal blue. The probably disordered regions (“loops”) within CagL which correspond to predicted loop regions in a CagL structural prediction (not shown) are indicated and numbered as loop 1 through loop 4 in B (also indicated as loops with the same numbering in Supplementary Fig. S2). The TSPSA motif was not structurally resolved in the crystal (3D) structure pdb: 3zcj_chainA and therefore is not indicated in B (loop 1).
