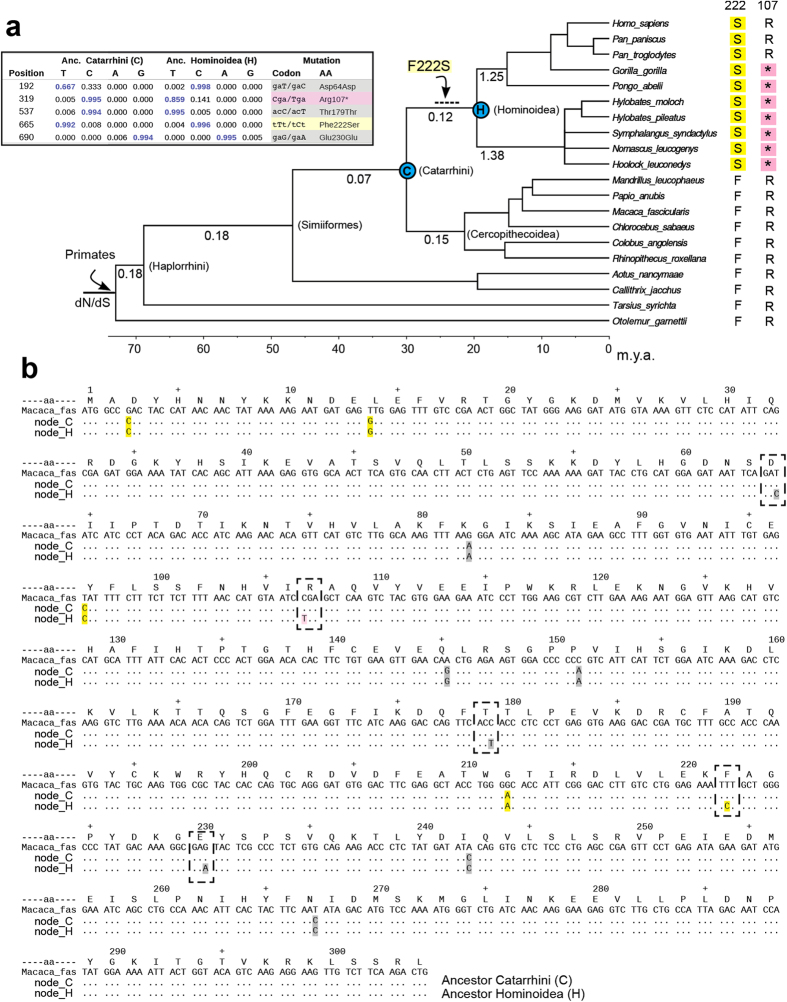
Figure 1. Phe→Ser mutation and the inactivation of the urate degradation pathway in hominoids.
(a) Chronogram of primate phylogeny based on multilocus analysis58; a polytomy was introduced to reflect uncertainties in the branching order of gibbon genera29,60. The tree was used as a framework for Bayesian reconstruction59 of ancestral Uox sequences and the ratio of non-synonymous/synonymous substitution rates (dN/dS). Posterior probabilities of nucleotide positions differing between the last common ancestors of Catarrhini and hominoids are shown in the inset. The single missense substitution in the hominoid lineage is indicated on the relevant branch (positions are according with the M. fascicularis sequence); the dashed line indicates uncertainty in dating the mutation along the branch. (b) The reconstructed Uox sequence of the Cathatrrini last common ancestor (node_C) compared with the sequence of the hominoid ancestor (node_H) using the sequence of M. fascicularis as a reference. Synonymous (gray), missense (yellow), and nonsense mutations (pink) with respect to the reference sequence are shown. Mutations which occurred along the branch connecting the two ancestral nodes are framed.