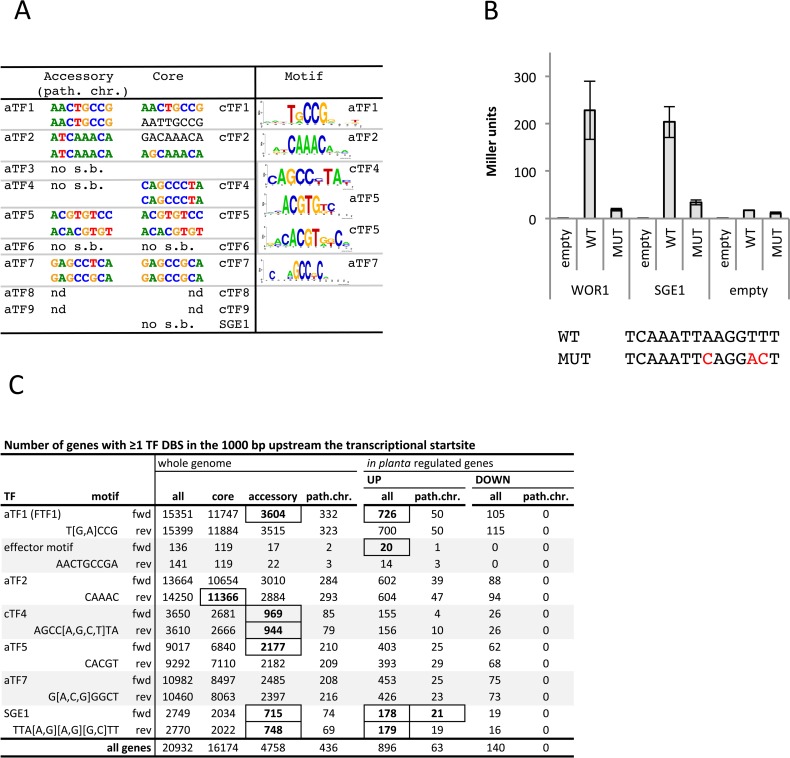
Fig 3. DNA binding motifs are not diversified within transcription factor families, but some motifs are enriched on accessory chromosomes.
A) DNA binding motifs determined by DNA binding arrays for several accessory and core transcription factor pairs. Accessory transcription factors were cloned from the pathogenicity chromosome in all cases except aTF1; this transcription factor was cloned from another accessory chromosome. Of transcription factors with a reliable DNA binding site on one or two arrays, the top 8-mers for both arrays are given (in color: significant enriched binding [E-score > 0.45], in grey, E-score < 0.45). For each transcription factor: upper 8-mer: HK array, lower 8-mer: ME array. For transcription factors with significantly enriched binding to ten 8-mers or more, a motif was constructed of all 8-mers with significant binding (right hand side of the figure). B) Sge1 can transcriptionally activate an UAS-less CYC1 promoter via the presence of the Wor1 DNA binding site. Activation is lost when the Wor1 DNA binding site is absent or mutated (lower panel). Activation assays were performed in duplicate, error bars represent standard deviation. C) The number of genes with one or more transcription factor DNA binding site (TF DBS) in the promoter, for the complete genome, for a subgenome (core, accessory or pathogenicity chromosome), and for a subset of genes (up- or down-regulated during infection). Boxes indicate a significant enrichment (P value < 0.01 after Bonferroni correction). The motif found in effector promoters earlier (aacTGCCGa) and overlapping with the Tf1 DNA binding site has been included.