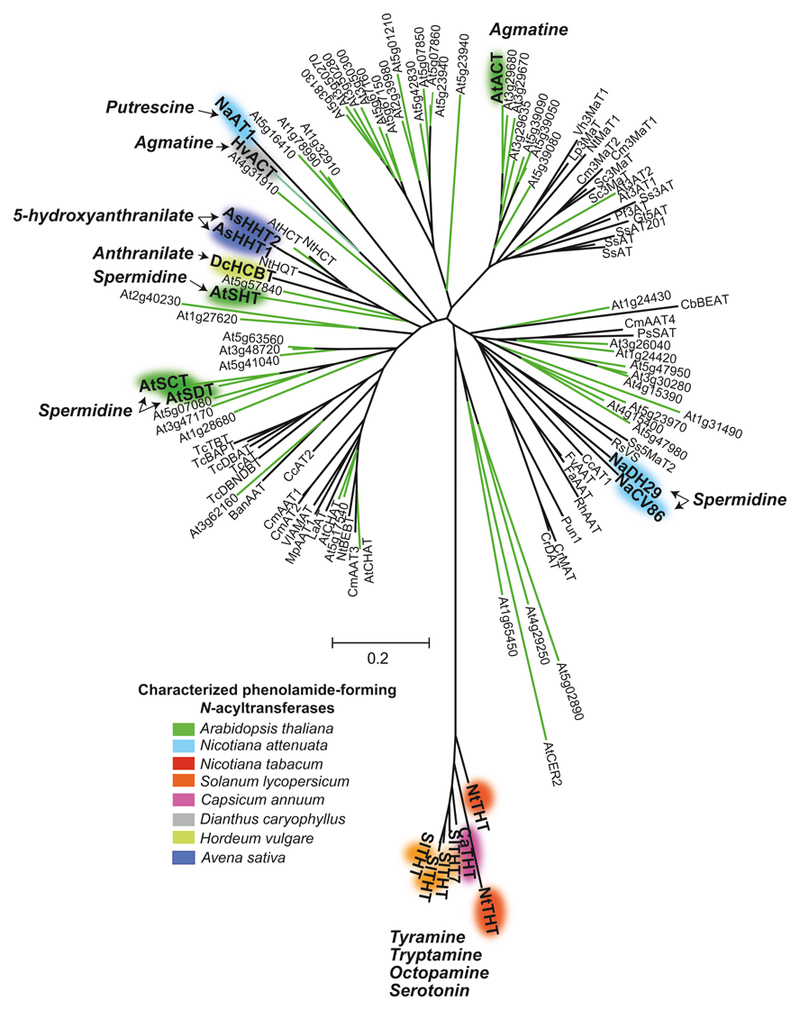
Figure 2. Phylogenetic relationships among phenolamide-forming N-acyltransferases.
A phylogenetic analysis was conducted for Arabidopsis BAHD (green branches of the tree) and functionally characterized N-acyltransferases including phenolamide-forming ones (acyl acceptor indicated) summarized in Bassard et al. (2010) and additional characterized N-acyltransferases reported in Luo et al. (2009). The phylogenetic tree reveals that N. attenuata DH29 and CV86 (highlighted in blue), which control the two-step synthesis of diacylated spermidines, cluster far from the Arabidopsis polyacylated spermidine-forming N-acyltransferases (AtSCT and AtSHT). Phenolamide N-acyltransferases with different acyl acceptor specificities (indicated next N-acyltransferases) are located on different parts of the tree. Sequences were aligned with Muscle and the alignment was trimmed with Gblocks to obtain 133 positions in 16 blocks that were used to calculate the phylogenetic tree using MEGA 4 and the Neighbor-Joining clustering method with 1000 iterations to calculate bootstrap values (Onkokesung et al. 2012). Colored ellipses of the tree connected to gene name in bold -- plant species names are reported in the color key -- denote for characterized phenolamide-forming N-acyltransferases. Plant species names are abbreviated as follows: Arabidopsis thaliana, At; Avena sativa, As; Capsicum annuum, Ca; Catharanthus roseus, Cr; Clarkia breweri, Cb; Curcumis melo, Cm; Dianthus caryophyllus, Dc; Fragaria anassa, Fa; Hordeum vulgare, Hv; Lupinus albus, La; Malus pumila, Mp; Nicotiana attenuata, Na; Nicotiana tabacum, Nt; Papaver somniferum, Ps; Salvia splendens, Ss; Solanum lycopersicum, Sl; Taxum cupsidata, Tc; Vitis labrusca, Vl.