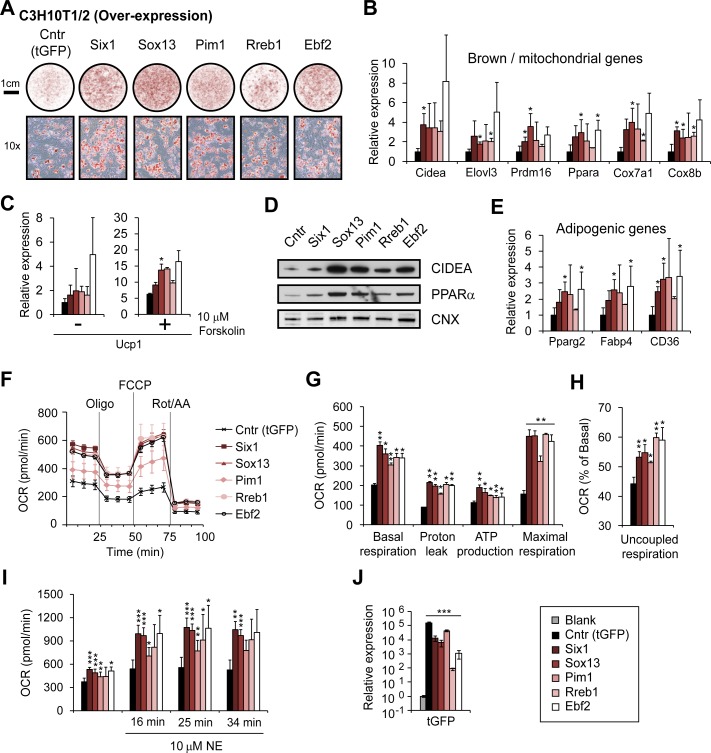
Fig 5. Over-expression of Six1, Sox13, Pim1 and Rreb1 promotes BA differentiation and function.
(A) Oil-Red-O staining of C3H10T1/2 cells differentiated for 7 days without BMP7 treatment. Cells were lenti-virally transduced to over-express the indicated genes tagged with tGFP before differentiation. Cells over-expressing tGFP only or the known brown adipogenic regulator Ebf2 served as negative and positive controls, respectively. (B) Expression of brown / mitochondrial marker genes was measured by qRT-PCR on day 7 of differentiation and found to be up-regulated upon over-expression of Six1, Sox13, Pim1, Rreb1, and Ebf2. (C) Expression of Ucp1 gene in cells treated with or without forskolin. (D) Protein levels of CIDEA and PPARα were examined by Western blot at day 7 of differentiation. Calnexin (CNX) was used as a loading control. (E) Expression of general adipogenic marker genes on day 7 of adipogenesis. (F) Oxygen consumption rates (OCR) measured in mature BAs over-expressing the indicated genes. (G) Basal respiration, proton leak, ATP production and maximal respiration were determined according to the OCR values in (F). (H) Uncoupled respiration was enhanced by over-expression of Six1, Sox13, Pim1, Rreb1, and Ebf2. (I) Over-expression of the indicated genes enhanced the response to Norepinephrine (NE) treatment in mature BAs. (J) qRT-PCR analysis confirmed the over-expression of the transduced genes using a primer pair targeting the tGFP coding sequence. Data in panel (B), (C), (E) and (J) represent the average of three independent experiments. Panel (F)—(I) show the representative result of two independent biological replicates (assayed in quadruplets). Error bars represent standard deviation. p-values (paired student’s t-test): * <0.05; ** <0.01; *** <0.001