Fig 5. Superposition of GtgE (cyan) and papain (magenta). Papain structure used for the comparison has PDB code 3CVZ.
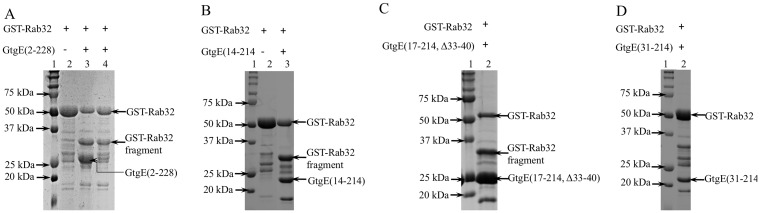
(A) Cleavage of GST-Rab32 by GtgE(2–228). Lane 1: protein markers, lane 2: GST-Rab32 fraction. The protein was only ~80% pure, other unidentified lower molecular weight bands were present in small amounts, lane 3: Cleavage of GST-Rab32 by GtgE(2–228) mixed in 2:1 (substrate:enzyme) molar ratio, lane 4: As lane 2 but 20:1 ratio of Rab32:GtgE(2–228). (B) Cleavage of GST-Rab32 by GtgE(14–214), lane 1: protein markers, lane 2: partially purified GST-Rab32 fraction, lane 3: Cleavage of GST-Rab32 by GtgE(14–228) in 2:1 molar ratio. (C) Cleavage of GST-Rab32 by GtgE(17–214, Δ33–40). Lane 1: protein marker,s lane 2: Cleavage of GST-Rab32 by GtgE(17–214, Δ33–40) in 2:1 molar ratio; D) GST-Rab32 in the presence fo GtgE(31–214). Lane 1: protein marker, lane 2: GST-Rab32 and GtgE(31–214) in 2:1 molar ratio. The unmarked bands were present in partially purified GST-Rab32 (see panel B lane 2), no band corresponding to the GST-Rab32 degradation product was observed.