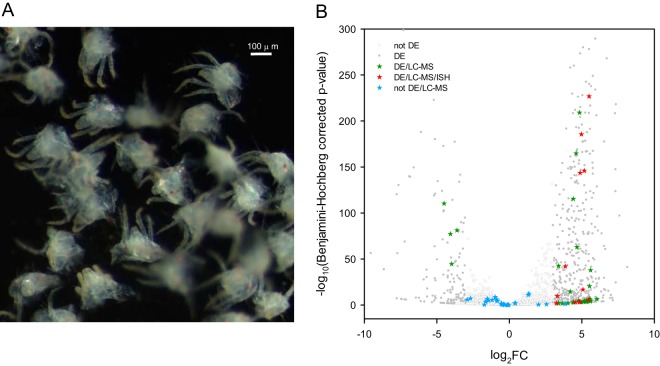
Fig. 2.
T. urticae genes that code for extracellular proteins and are differentially expressed between proterosomas and intact females. A, Dissected T. urticae proterosomas used for RNA extraction (see Fig. 3 for dissection position). B, The negative log10 of Benjamini-Hochberg adjusted p values was plotted against the log2FC in gene expression for all T. urticae genes (see supplemental Fig. S2) and subsequently filtered for genes coding for extracellular proteins (predicted to be extracellular by WoLF PSORT (82) and predicted with a SP by SignalP 4.1(81), see supplemental Table S9). Differentially expressed genes (|FC| ≥ 8, Benjamini-Hochberg adjusted p value ≤ 0.05) are shown as gray dots (DE), whereas genes that were not differentially expressed are shown as gray circles (not DE). Genes coding for extracellular proteins that were identified by nano-LC-MS/MS (“extracellular LC-MS salivary protein genes”) are depicted as stars. Extracellular LC-MS salivary protein genes that were differentially expressed are depicted as green stars (DE/LC-MS). Extracellular LC-MS salivary protein genes that were differentially expressed and for which ISH confirmed expression in salivary glands are depicted as red stars (DE/LC-MS/ISH) whereas extracellular LC-MS salivary protein genes that are not differentially expressed are shown as blue stars (not DE/LC-MS).