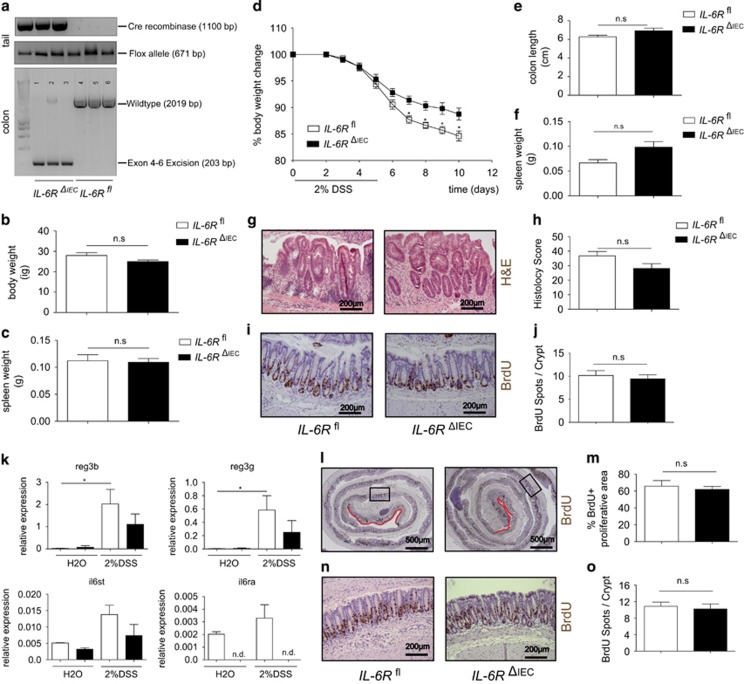
Figure 3.
IL-6RΔIEC are not susceptible to DSS colitis and display regular epithelial regeneration. (a) Genotyping of IL-6Rfl and IL-6RΔIEC, showing expression of Cre recombinase and the mutant IL-6R allele in tail PCR. IEC-specific deletion of IL-6R was verified by isolating colon DNA and using primer pairs spanning the exon 4–6 region of the IL-6R gene (il6ra). PCR products show an amplicon of 2016 bp in IL-6Rfl and a truncated amplicon of 203 bp in IL-6RΔIECplus a faint 2016 bp signal, indicating DNA from non-epithelial origin. (b, c) Body weight and spleen weight of sex-matched untreated IL-6Rf(n=4) mice and IL-6RΔIEC (n=4) at the age of 8–12 weeks. (d) Sex-matched littermate IL-6Rfl (n=8) and IL-6RΔIEC (n=18) mice aged 8–12 weeks were subjected to experimental colitis induced by adding 2% of DSS to the drinking water. Body weight changes relative to the starting weight on day 0 showed that IL-6RΔIEC mice lost less weight than their IL-6Rfl littermates. (e, f) Postmortem analyses showed no significant difference between IL-6RΔIEC and IL-6Rfl in colonic shortening (e) or relative spleen weight (f). (g, h) H&E staining of colon Swiss rolls (g) and corresponding histological disease scores42 (h) showed no significant differences in disease severity between IL-6RΔIEC and IL-6Rflmice. (i, j) Representative pictures of BrdU staining of IL-6RΔIEC and IL-6Rfl are shown in (i). 10 mg/kg bodyweight of BrdU was injected i.p. 1.5 h before sacrifice. A minimum of 15 crypts/intestine was counted and genotype pooled data from IL-6Rfl (n=6) and IL-6RΔIEC (n=12) are depicted for statistical evaluation. No significant difference was seen in the number of BrdU+ cells/crypt between IL-6RΔIEC and IL-6Rfl mice (j). (k) IEC-specific expression of STAT3 target genes. Mice (n>3 per genotype) were challenged with 2% DSS or left untreated (water, H2O) for 3 days, and colonic IECs were isolated. Gene expression was assessed using qPCR. DSS induction led to significant upregulation of STAT3 target genes in IL-6Rfl animals, which was reduced or abrogated in IL-6RΔIEC mice. (l, m) Representative pictures of BrdU staining of IL-6RΔIEC and IL-6Rfl are shown in (i). 10 mg/kg bodyweight of BrdU was injected i.p. 1.5 h before sacrifice. The percentage of intact BrdU+ mucosa was expressed as 1−(length of BrdU-negative mucosa in μm/total mucosal length in μm)*100. (n, o) A minimum of 15 crypts/intestine were evaluated for BrdU-positive stained cells. Genotype pooled data from IL-6Rfl (n=4) and IL-6RΔIEC (n=4) are depicted for statistical evaluation. Significance was determined using the two-tailed Student's t-test, and data are expressed as mean±s.d. *P<0.05.