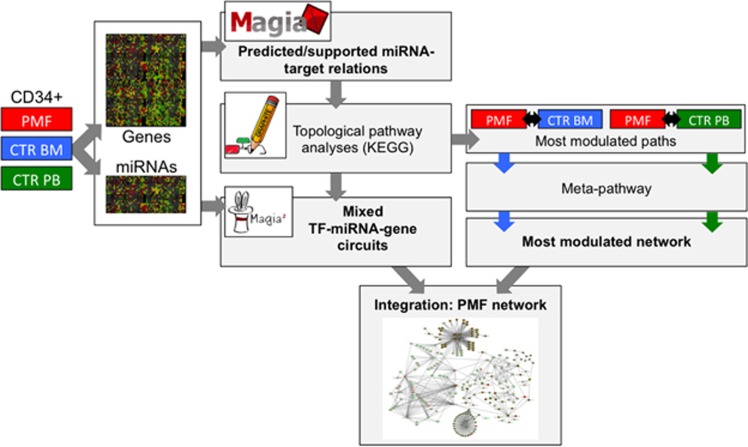
Figure 1.
Experimental design and analysis procedures flow chart. miRNA and gene expression data in CD34+ cells of PMF patients and healthy controls were considered; CTR BM and CTR PB controls were considered separately in parallel analyses, and results were merged at the final steps. MAGIA integrated analysis outputted the subset of predicted miRNA-target relations supported by expression data; these were used to enrich KEGG pathway-derived miRNA-gene networks based on pathways annotations and on validated miRNA-target relations. Topological pathway analyses by Micrographite identified most modulated paths, showing significant gene/miRNA expression variations and changes in relations strength, then a non-redundant comprehensive meta-pathway was derived; iterative analysis of the meta-pathway identified the most modulated network for each comparison. Magia2 analysis identified mixed TF-miRNA-gene circuits. Results of parallel comparisons were merged in an integrated network.