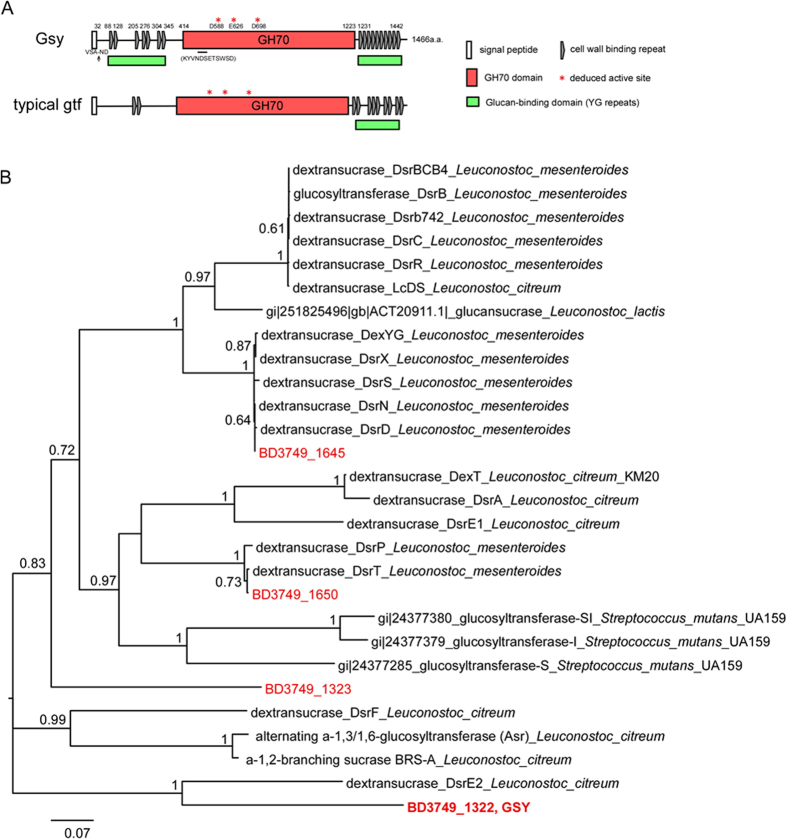
Figure 5. Sequence-based analysis of Gsy.
(A) Domain structure analysis of Gsy. The deduced active sites are marked with asterisks, and the peptide used for antibody production is indicated below the GH70 domain. Cell wall binding repeats were analyzed on Scanprosite (http://prosite.expasy.org/scanprosite). Shown below Gsy is a typical Gtf from S.mutans (AAN58706.1)(http://www.cazy.org/GH70_characterized.html, http://prosite.expasy.org/scanprosite/). (B) Phylogenetic analysis of putative glucansucrases in BD3749 and the characterized GH70 enzymes. The tree was constructed with the maximum-likelihood method based on the JTT matrix-based model using MEGA657. Each protein is labelled with its GenBank accession number. GH70 enzymes of BD3749 are labelled in red. The value on each branch is the estimated confidence limit for the position of the branches, as determined by bootstrap analysis. The scale bar represents a 7% difference in amino acid sequence58. It should be noted that CDX66820.1 and CAB65910.2, GH70 enzymes from NRRL B-1299 and NRRL B-1355, respectively, were assigned to Leuconostoc citreum as suggested by Bounaix et al.59.