Abstract
BRCA1 and BRCA2 are tumour-suppressor genes encoding proteins that are essential for the repair of DNA double-strand breaks by homologous recombination (HR). Cells that lack either BRCA1 or BRCA2 repair these lesions by alternative, more error-prone mechanisms. Individuals carrying germline pathogenic mutations in BRCA1 or BRCA2 are at highly elevated risk of developing breast and/or ovarian cancer. Genetic testing for germline pathogenic mutations in BRCA1 and BRCA2 has proved to be a valuable tool for determining eligibility for cancer screening and prevention programmes. In view of increasing evidence that the HR DNA repair pathway can also be disrupted by sequence variants in other genes, screening for other BRCA-like defects has potential implications for patient care. Additionally, there is a growing argument for directly testing tumours for pathogenic mutations in BRCA1, BRCA2 and other genes involved in HR-DNA repair as inactivation of these genes may be strictly somatic. Tumours in which HR-DNA repair is altered are most likely to respond to emerging targeted therapies, such as inhibitors of poly-ADP ribose polymerase. This review highlights the biological role of pathogenic BRCA mutations and other associated defects in DNA damage repair mechanisms in breast and ovarian cancer, with particular focus on implications for patient management strategies.
Introduction
BRCA1 and BRCA2 are tumour-suppressor genes located on chromosomes 17q21 and 13q12, respectively.1, 2, 3, 4 Functional BRCA proteins are involved in the maintenance of genome stability through repair of DNA double-strand breaks (DSBs) by homologous recombination (HR), cell growth regulation and control of cell division.5, 6 Individuals carrying monoallelic germline pathogenic mutations in BRCA1 or BRCA2 (BRCA1/2) are at higher risk of developing a variety of cancers, particularly breast and/or ovarian cancer. Meta-analyses have indicated a mean cumulative breast cancer risk at age 70 years to be 57% (95% CI 47–66%) for patients carrying the BRCA1 pathogenic mutations and 49% (95% CI 40–57%) for patients carrying the BRCA2 pathogenic mutations.7 The equivalent mean cumulative ovarian cancer risk is 40% (95% CI 35–46%) for patients carrying the BRCA1 pathogenic mutations and 18% (95% CI 13–23%) for patients carrying the BRCA2 pathogenic mutations.7 A prospective epidemiological study (EMBRACE) showed that carriers of BRCA1 and BRCA2 pathogenic mutations have a mean cumulative risk of breast cancer at age 70 years of 60% (95% CI 44–75%) and 55% (95% CI 41–70%), respectively.8 The equivalent mean cumulative ovarian cancer risk is 59% (95% CI 43–76%) and 16.5% (95% CI 7.5–34%), respectively.8 Tumourigenesis in germline BRCA1/2 pathogenic mutation carriers generally follows a two-hit hypothesis, the first ‘hit' owing to the inherited pathogenic mutation of one BRCA allele and the second ‘hit' owing to the somatic inactivation of the second-wild-type allele.9, 10, 11 Increasing evidence suggests that other types of breast and ovarian cancers share genomic and phenotypic similarities with tumours associated with germline and somatic BRCA1/2 pathogenic mutations.12 Such cases may be sensitive to the same emerging targeted therapies as tumours associated with germline BRCA1/2 pathogenic mutations.
Methods for the detection of BRCA1 and BRCA2 pathogenic mutations are now widely accessible. Until now, the principal aim of BRCA1/2 pathogenic mutation testing has been to enable risk assessment to permit early diagnosis and cancer prevention. However, it is increasingly apparent that knowledge of BRCA status has prognostic utility that can affect treatment decisions and may improve survival.13, 14, 15 This review highlights the biological role of BRCA1/2 pathogenic mutations and other associated defects in DNA damage repair in breast and ovarian cancer, with particular focus on implications for clinical management strategies.
BRCA and repair of DNA DSBs by HR
DNA repair is essential for the survival of both normal and cancer cells. DNA repair mechanisms also allow cancer cells to survive the DNA injury imposed by chemotherapy or radiation. An elaborate network of genome surveillance systems and DNA repair mechanisms exist to repair DNA lesions and ensure the integrity of the genome and hence cell fitness and viability. DNA DSBs, in which both strands of the double helix are severed, are the most dangerous type of DNA lesion; if left unrepaired, or repaired incorrectly, DSBs may result in massive loss of genetic information, genomic rearrangements or cell death. Two different mechanisms exist for the repair of DSBs: non-homologous end joining (NHEJ) and HR.16 These pathways differ in their fidelity and template requirements. NHEJ is an intrinsically error-prone pathway, which modifies the broken DNA ends, and ligates them together with little or no homology, generating small deletions or insertions. In contrast, HR is a highly conserved pathway that provides accurate repair of DSBs in the late S and G2 phases of the cell cycle using the intact sister chromatid as a template to repair the break and maintain sequence integrity.
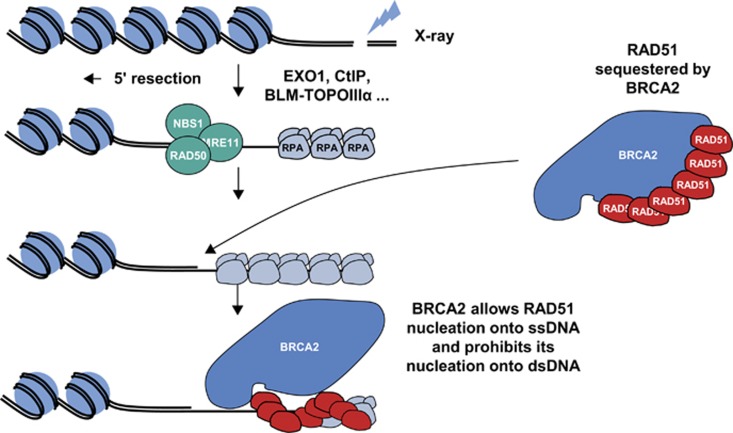
BRCA1 and BRCA2 are key components of the HR pathway, and cells that lack these proteins are unable to repair DSBs by HR.6, 9 BRCA1 appears to have an early and broad role in the promotion and regulation of HR.5, 9, 17, 18 BRCA1 has been shown to colocalise at sites of DNA damage with RAD51, another key protein involved in HR, while BRCA1/2-deficient cell lines lack RAD51 foci.19 BRCA1 appears to regulate HR, at least in part, through a modulatory role in the PALB2-dependent loading of BRCA2-RAD51 repair machinery at DNA breaks. A central role for BRCA2 in HR was first suggested by evidence showing the acquired chromosomal abnormalities of BRCA2-deficient cell lines to be similar to those seen in Fanconi's anaemia.18, 20, 21 Furthermore, cell lines derived from some patients with Fanconi's anaemia were shown to carry biallelic pathogenic mutations in BRCA2, which led to BRCA2 also being referred to as FANCD1.20, 22 BRCA2 knockout cells sustain spontaneous aberrations in chromosome structure that accumulate during division in culture.21 In the absence of DNA damage, RAD51 is sequestered by BRCA2, prohibiting RAD51 nucleation onto double-stranded DNA (Figure 1). Following DNA damage, BRCA2 relocalises to the site of DNA damage and enables RAD51 nucleation onto single-stranded DNA.23, 24
Figure 1.
Role of BRCA2 in DNA DSB repair by HR. Diagram courtesy of Gaël Millot, Curie Institute and University Pierre and Marie Curie, Paris, France.23, 24
Other causes of defects in HR
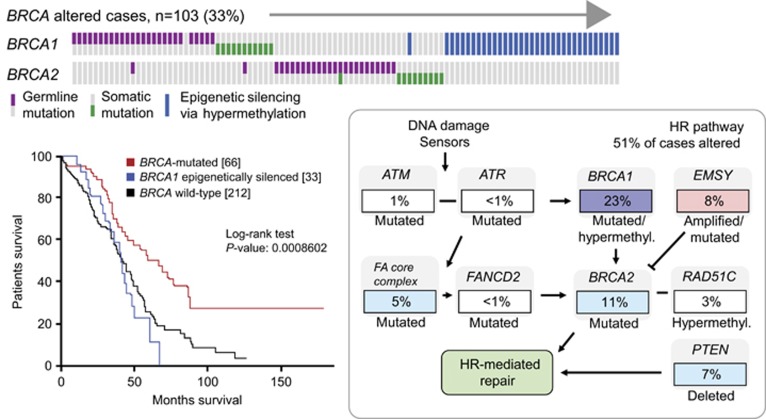
Pathogenic mutations in several other genes involved in HR-mediated DNA repair have been shown to be associated with breast and ovarian cancer predisposition (Figures 2 and 3).25, 26 In the Cancer Genome Atlas Research Network analysis of high-grade serous ovarian adenocarcinomas, genomic alterations in 26% of HR genes other than BRCA1/2 were observed, including amplification or pathogenic mutation of EMSY (8%), promoter methylation of RAD51C (3%), pathogenic mutation of ATM/ATR (2%) and pathogenic mutation of Fanconi's anaemia genes (5%) (Figure 2).25 In addition, focal deletion or pathogenic mutation of PTEN (7%) has been observed; however, the role of PTEN in HR or as a surrogate of HR deficiency remains to be determined. Such cancers are said to display ‘BRCAness' if they exhibit similar DNA repair defects to those seen in BRCA1/2-deficient cells.27
Figure 2.
High-grade ovarian carcinoma: frequency of BRCA1/2 inactivation in and impact on survival; frequency of alterations of genes involved in DNA damage sensors or HR (and not repair), including BRCA1 and BRCA2.25 Reprinted by permission from Macmillan Publishers Ltd, copyright 2011.
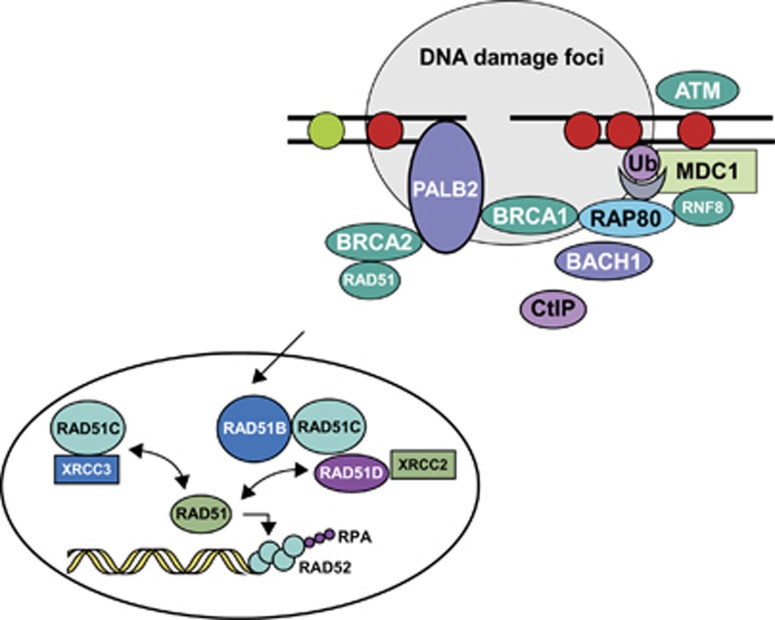
Figure 3.
Proteins involved in DNA DSB repair by HR. RPA, replication protein A. Figure adapted from Sy et al.26 and Skorski.39 Reprinted by permission from Macmillan Publishers Ltd, copyright 2002, and from PNAS.
Monoallelic germline pathogenic mutations in PALB2, BRIP1 and ATM have been shown to be associated with an increased breast cancer relative risk of approximately 2–3.28, 29, 30, 31, 32 Biallelic pathogenic mutations in PALB2 and BRIP1 have been observed in patients with Fanconi's anaemia, resulting in their alternative names of FANCN and FANCJ, respectively.33, 34 More recently, BRIP1 pathogenic mutations have also been demonstrated in patients with ovarian cancer.35 The frequency of germline PALB2 pathogenic mutations in ovarian cancer cases does not appear increased as compared with the general population.36 PALB2 encodes the partner and localiser of BRCA2 protein, which stabilises the BRCA2 protein and anchors it to structures within the nucleus, allowing the BRCA2 protein to mediate DNA repair.26 BRIP1 – also known as BACH1 – encodes BRCA1-interacting protein-terminal helicase 1, a DNA helicase that influences the DNA repair ability and tumour-suppressor function of BRCA1.37 ATM is a protein kinase with a key role in sensing DNA DSBs and monitoring their repair. Biallelic germline inactivation of ATM is responsible for the neurodegenerative disorder ataxia-telangiectasia.38
The RAD51 paralogues, RAD51C and RAD51D, also have an integral role in the repair of DNA DSBs through HR (Figure 3).39 To date, germline pathogenic mutations in RAD51 have not been observed in patients with breast or ovarian cancer, suggesting that they are lethal. However, RAD51C pathogenic mutations have been identified in up to 2.9% of highly penetrant breast and ovarian cancer families who previously screened negative for BRCA1/2 pathogenic mutations.40, 41, 42 Ovarian cancer occurrence in families with RAD51C pathogenic mutations shows major similarities with families carrying BRCA1/2 pathogenic mutations. In addition, as these families show apparent segregation of the pathogenic mutation with the cancer phenotype, the penetrance of RAD51C pathogenic mutations is predicted to be comparable to that of BRCA2 pathogenic mutations (Antoniou, unpublished data). However, the mean age at onset for ovarian cancer observed in women with RAD51C pathogenic mutations is approximately 60 years, which is older than in BRCA1 pathogenic mutation carriers (51 years).43 The paralogues RAD51B, RAD51D and XRCC2 are also associated with an increased ovarian cancer risk.44, 45, 46, 47, 48 Additional studies are required to better define their contribution to breast and ovarian cancer predisposition.
Synthetic lethality: a potential therapeutic approach
Two genes are said to be synthetically lethal if a mutation in either gene alone is compatible with cell viability but simultaneous mutation of both genes causes cell death.49 This suggests that inhibition of an additional DNA damage repair pathway is likely to be synthetically lethal in cells lacking HR, whether through germline or somatic BRCA1/2 pathogenic mutations, posttranslational changes of BRCA or abnormalities of other genes involved in HR. The base excision repair pathway, in which poly-ADP ribose polymerase (PARP) has a major role, is important for the repair of certain kinds of DNA damage, particularly in the absence of HR. Loss of PARP function results in the accumulation of single-strand DNA breaks, which are subsequently converted to DSBs by cellular transcription and replication. These DSBs, which are typically repaired by HR or NHEJ in normal cells, would accumulate in HR-deficient cells, leading to subsequent cell death.
Loss of PARP1 function induces the formation of nuclear RAD51 foci as a result of the increased formation of DNA lesions that need to be repaired by the HR pathway. Two pivotal preclinical studies demonstrated loss of these RAD51 foci in BRCA1- and BRCA2-deficient cells after PARP inhibitor exposure.50, 51 Sensitivity to PARP inhibition has also been observed in cells with defects in HR other than BRCA deficiency.52 Several PARP inhibitors are now in various stages of clinical development.
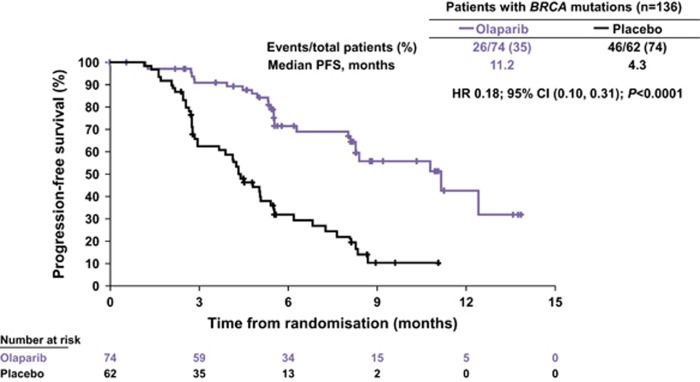
Olaparib is a potent oral PARP inhibitor that has been shown to induce synthetic lethality in BRCA1/2-deficient tumour cells.53, 54 Antitumour activity of olaparib has been demonstrated in patients with BRCA-mutated breast and ovarian cancer in proof-of-concept trials.55, 56 In a Phase II trial, maintenance therapy with olaparib was shown to significantly prolong progression-free survival in patients with platinum-sensitive relapsed serous ovarian cancer compared with placebo, with the greatest benefit seen in patients with a pathogenic BRCA mutation.57 An 82% reduction in risk of disease progression with olaparib compared with placebo was seen in patients with pathogenic BRCA mutations (Figure 4). Retrospective exploratory analyses of time to subsequent treatment or death and time to second subsequent treatment or death (indicators of the postprogression efficacy of olaparib) also showed significant advantages in favour of olaparib over placebo in both the overall population and in patients with pathogenic BRCA mutations. Further studies of olaparib and other PARP inhibitors in this setting are ongoing.
Figure 4.
Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer: progression-free survival in patients harbouring a pathogenic BRCA mutation.57 Reprinted from Ledermann et al.,57 copyright 2014, with permission from Elsevier.
In addition to potential utility for the treatment of tumours harbouring a pathogenic BRCA1/2 mutation, PARP inhibition might also be a useful therapeutic approach for the treatment of a wider range of tumours bearing a variety of deficiencies in the HR pathway and thus displaying properties of 'BRCAness'. Methods to identify patients most likely to benefit from these emerging therapies are therefore required.
Moving from cancer prevention to cancer treatment: a paradigm change for BRCA1/2 testing process?
At present, the primary goal of genetic testing for pathogenic mutations in BRCA1 and BRCA2 is identification of women at the greatest risk of developing breast and ovarian cancer to enable appropriate preventative strategies to be implemented. However, now and in the near future, testing for pathogenic BRCA mutations or BRCA-like defects is likely to have a key role in patient care; for example, in the identification of patients who are most likely to benefit from emerging therapies targeting DNA repair mechanisms, such as PARP inhibitors.
In unselected breast cancer, the reported pathogenic BRCA mutation frequency is 1–5%, whereas in unselected ovarian cancer the reported frequency is higher at 6–16%.13, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 It should be noted that these ranges exclude studies in male breast cancer and Ashkenazi Jewish women. Higher prevalence is associated with factors such as a positive family history, young age at onset, male breast cancer and multiple tumours in the same patient or certain histological characteristics.58, 74
In an Australian population-based, case–control study, 1001 women with newly diagnosed histologically confirmed, non-mucinous, invasive epithelial ovarian, peritoneal or fallopian tube cancer were screened for point mutations and large rearrangements in BRCA1 and BRCA2.13 Pathogenic BRCA1/2 mutations were identified in 14.1% of patients (141 mutations in 1001 women; 95% confidence interval 11.9, 16.3). Of these 141 pathogenic mutations, 88 were in BRCA1 (62.4%) and 53 were in BRCA2 (37.6%). BRCA1/2 pathogenic mutations were seen in 16.6% of serous tumours, 16.8% of high-grade tumours and 17.1% of all high-grade serous tumours. Of note, 44% of women with BRCA1/2 germline pathogenic mutations in this study had no previous family risk factors.
In a study of 489 high-grade serous ovarian adenocarcinomas, 64/316 (20.3%) tumours with sequenced exomes (n=316) were found to harbour pathogenic BRCA1 or BRCA2 mutations; two tumours were observed to harbour both BRCA1 and BRCA2 mutations.25 The majority of detected BRCA1/2 pathogenic mutations were germline in origin (71%), corresponding to a germline pathogenic variant frequency of 14.6% and a somatic pathogenic variant frequency of 6.0%. Accompanying heterozygous loss was observed with 81 and 72% of BRCA1 and BRCA2 pathogenic mutations, respectively, indicating inactivation of both alleles, as predicted by Knudson's two-hit hypothesis for a tumour-suppressor gene. In addition to BRCA1/2 pathogenic mutations, 34/316 (10.8%) of tumours had lost BRCA1 expression through BRCA1 promoter methylation.25 Epigenetic silencing of BRCA1 was shown to be mutually exclusive of germline BRCA1/2 pathogenic mutations (Figure 2). Survival analysis based on BRCA status revealed divergent outcomes, with longer overall survival for BRCA-mutated cases compared with BRCA wild-type and BRCA1 epigenetically silenced cases, respectively (Figure 2).25 These data would suggest that a broader range of patients with ovarian cancer should now be tested, perhaps offering BRCA testing to all but patients whose tumours have mucinous histology.
So far, the low frequency of heterozygotes for BRCA pathogenic mutations relative to the high incidence of breast and ovarian cancer, coupled with the high costs of BRCA testing, has rendered exhaustive testing of all patients with breast and ovarian cancer impractical. However, the evolution of multiple gene panel sequencing and the decreasing cost of genetic testing are allowing health-care budgets to offer pathogenic BRCA mutation testing to broader populations without a significant impact on cost.
Pathogenic BRCA mutation testing for cancer prevention
The estimated population frequency of pathogenic BRCA1/2 mutations is 1:800–1:1000 per gene; however, the prevalence of BRCA1/2 germline pathogenic mutations varies considerably between different ethnic groups and geographical areas.74 BRCA1 and BRCA2 pathogenic mutation frequency in patients with breast and ovarian cancer unselected for family history or age at onset is generally low.74 At present, the selection of appropriate candidates for testing is typically based on country-specific guidelines or by larger international societies.74 Widely accepted clinical criteria for referral include family history and age at cancer onset. Based on these criteria, pathogenic mutations in BRCA1 or BRCA2 are typically identified in 12–15% of tested cases.
Guidelines in some countries require a 10–20% probability of detecting a BRCA1/2 pathogenic mutation within a family before mutational analysis is considered. A number of predictive models and scoring systems have been developed to assess the probability of a pathogenic BRCA1/2 mutation in a given individual dependent on their family history, with varying degrees of validation.75 These include BRCAPRO, BOADICEA or the Manchester Scoring System.74 Methods that compute carriage probabilities are the most predictive but require specific computer software and data entry for all family members can be time-consuming. Scoring systems avoid data entry, are a good proxy to probability computations and can generally be easily modified to include new relevant predictive factors.76
The Manchester scoring system allocates a score to each affected individual in a family and computes the sum of the scores in the maternal and paternal lineage to determine whether or not a genetic test is recommended.77, 78 The Manchester scoring system is empirical as the scores have been determined to fit the observations made in a group of selected families tested for pathogenic BRCA1/2 mutations. It does not take into account information on unaffected family members, the presence of which is expected to decrease the probability of mutations, and gives the same weight to all affected family members whatever their degree of kinship.
An alternative system has been recently developed, which has been shown to be superior to the Manchester scoring system.76 The new scoring system is based on the conditional probability P that a proband is a carrier, given all relevant predictive information in the family. Parameters taken into account include pathogenic BRCA1/2 mutation frequencies in the population, as well as breast and ovarian cancer risks in carriers and non-carriers. The performance of the new scoring system was evaluated using a simulation of 10 million families, built from women affected with breast or ovarian cancer at different ages.76, 79 At a score threshold of 5, where positive predicted value (PPV) (15% ie, percentage of carriers among tested individuals) and specificity (87% ie, percentage of non-tested individuals among non-carriers) are similar to the Manchester scoring system with a pathogenic mutation probability of 10%, sensitivity (ie, percentage of tested individuals among carriers) with the new scoring system was higher than the Manchester scoring system (77 vs 72%).76 The improved performance of the new scoring system compared with the Manchester scoring system was attributed to accounting for unaffected family members and for the degree of kinship of relatives with the proband.
Pathogenic BRCA mutation testing for cancer treatment
Pathogenic BRCA mutation testing on a broader population is likely to result in increased numbers of relatives screened, which may subsequently reduce the future incidence of ovarian cancer, as those relatives who are shown to harbour a pathogenic BRCA mutation can be started on a cancer prevention programme.
Despite mutation detection threshold (PPV) lowering,76 there remains a need to broaden BRCA1/2 testing criteria in order to optimise use of BRCA testing to guide treatment decisions. One approach to broadening BRCA1/2 testing criteria is by introducing individual criteria, such as including women with triple-negative (TN) breast cancer (ie, negative for oestrogen receptor, progesterone receptor expression and HER2 amplification) and women with high-grade ovarian cystoadenocarcinoma.
In one recent analysis,79 probabilities for the TN status of a breast tumour were obtained from the proportion of TN tumours among women tested for BRCA1/2 in studies without any selection on morphological characteristics.80, 81, 82, 83 A Bayesian model was developed to calculate the probability of a pathogenic BRCA1 mutation according to age at diagnosis, assuming the rate of TN disease to be 68% among women with a pathogenic BRCA1 mutation and 13% among women with no pathogenic BRCA1 mutation (including those with a pathogenic BRCA2 mutation). Results showed the probability of a pathogenic BRCA1 mutation to be high at 23% in women diagnosed with TN breast cancer at <35 years, compared with 9.2% in women diagnosed at 35–39 years, 5.5% at 40–49 years, 4.3% at 50–59 years and 2.5% at 60–69 years (unpublished data). Thus TN disease status appears to be a good marker for pathogenic BRCA1 mutations especially in young women with breast cancer.
Including individual criteria – specifically, ovarian cancer before age 61 years (except borderline, mucinous tumours), breast cancer before age 36 years, TN breast cancer before age 51 years and male breast cancer at any age – into one predictive model increased sensitivity to 77% (a gain of 13% compared with use of French family criteria alone), with slight reductions in PPV (11%) and specificity (82%).76, 79 However, the ‘cost' of introduction of individual criteria was to increase the number of tests by 49%.
In order to maximise the potential of emerging therapies, such as PARP inhibitors and agents targeting other proteins involved in DNA repair mechanisms, identification of patients with germline pathogenic mutations in other genes or biallelic somatic inactivation of genes involved in DNA repair is likely to become increasingly important. There is increasing interest in confirming the pathogenicity of many sequence variants in the BRCA1/2 genes whose pathogenicity is currently unknown, as well as other genes in the HR-pathway. Three groups have recently reported similar genomic ‘scars', which appear to be potential surrogate markers of HR deficiency and potential sensitivity to DNA-damaging agents. These include a HR deficiency score based on genome-wide loss of heterozygosity,84 a telomeric allelic imbalance score85 or large-scale genomic instability.86 The potential clinical application of genomic ‘scars' include facilitating the use of PARP inhibitors and platinum-based chemotherapy in breast, ovarian and other cancers;84 identifying cancer patients likely to benefit from treatments targeting defective DNA repair;85 and easing the challenge of selecting patients for genetic testing or recruitment to clinical trials of novel emerging therapies that target DNA repair deficiencies in cancer.86 Such genomic ‘scars' would appear to merit further study as potential biomarkers of response to emerging targeted therapies; however, direct testing of BRCA/HR-associated genes by Next-Generation Sequencing (NGS) without investigating for such markers may be more practical.
The evolution of genetic testing may allow a more accessible option for broader testing strategies. NGS is an efficient and cheaper alternative to sequential testing, allowing simultaneous screening of multiple cancer susceptibility genes. Assessing multiple gene panels will allow individualised testing dependent upon the risk for the individual from factors such as age, ethnicity or likelihood of inherited predisposition by family history of disease.87 In the future, it is anticipated that more panels of genes linked to HR defects and cancer risks will be available at a lower cost.87 In addition, the use of such gene panels directly on tumours in order to identify biallelic somatic events is an important issue and a technological challenge. It is of course important, whatever the strategy used, that patients are kept well informed of the testing process.
Conclusions
It is increasingly apparent that the HR pathway for DNA repair is not only disrupted by germline and somatic pathogenic BRCA1/2 mutations but also by pathogenic mutations in other genes involved in HR. The presence of BRCA-like defects in patients with breast and ovarian cancer can be used to inform clinical management decisions. As such, there is a clear need to broaden the criteria for BRCA1/2 germline genetic testing, as well as to expand testing to include identification of germline pathogenic mutations in other genes that may be involved in DNA damage repair by HR DSBR. Furthermore, tumour markers of ‘BRCAness' may have utility for the identification of patients most likely to respond to emerging targeted therapies, such as PARP inhibitors. With the rapid evolution of multi-panel genetic sequencing, coupled with the decreasing cost of genetic testing, BRCA testing and individualised screening is becoming more accessible.
Acknowledgments
This work was sponsored by AstraZeneca. Medical writing services were provided by Tom Hudson of iMed Comms, Macclesfield, UK and were funded by AstraZeneca.
Footnotes
Dominique Stoppa-Lyonnet has received payments for speaker services from AstraZeneca. Her laboratory is supported by public funds from the French National Cancer Institute and AstraZeneca France, Inserm-Transfert.
References
- Hall JM, Lee MK, Newman B et al: Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990; 250: 1684–1689. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D et al: A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266: 66–71. [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Rommens J et al: The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 1996; 12: 333–337. [DOI] [PubMed] [Google Scholar]
- Wooster R, Neuhausen SL, Mangion J et al: Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 1994; 265: 2088–2090. [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A: The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006; 25: 5864–5874. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR: Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci 2001; 114: 3591–3598. [DOI] [PubMed] [Google Scholar]
- Chen S, Parmigiani G: Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 2007; 25: 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Peock S, Frost D et al: Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 2013; 105: 812–822. [DOI] [PubMed] [Google Scholar]
- Collins N, McManus R, Wooster R et al: Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene 1995; 10: 1673–1675. [PubMed] [Google Scholar]
- Smith SA, Easton DF, Evans DG, Ponder BA: Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet 1992; 2: 128–131. [DOI] [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M et al: DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 2001; 10: 3001–3007. [DOI] [PubMed] [Google Scholar]
- Burgess M, Puhalla S: BRCA 1/2-mutation related and sporadic breast and ovarian cancers: more alike than different. Front Oncol 2014; 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop K, Fereday S, Meldrum C et al: BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012; 30: 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Hays JL, Annunziata CM et al: Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst 2014; 106: dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KP, Walsh T, Harrell MI et al: Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014; 20: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V: DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008; 7: 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt AN, Lord CJ, McCabe N et al: Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol 2005; 70: 139–148. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR: Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002; 108: 171–182. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A et al: Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 1997; 88: 265–275. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S et al: Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002; 297: 606–609. [DOI] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H et al: Involvement of Brca2 in DNA repair. Mol Cell 1998; 1: 347–357. [DOI] [PubMed] [Google Scholar]
- Patel KJ: Fanconi anemia and breast cancer susceptibility. Nat Genet 2007; 39: 142–143. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ: Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012; 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Jensen RB: BRCA2: one small step for DNA repair, one giant protein purified. Yale J Biol Med 2013; 86: 479–489. [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network: Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J: PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA 2009; 106: 7155–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A: Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer 2004; 4: 814–819. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Casadei S, Heikkinen T et al: Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Hart SN, Sharma P et al: Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 2015; 33: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D et al: PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 2007; 39: 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S et al: ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 2006; 38: 873–875. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A et al: Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 2006; 38: 1239–1241. [DOI] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC et al: The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet 2005; 37: 934–935. [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H et al: Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 2007; 39: 162–164. [DOI] [PubMed] [Google Scholar]
- Kanchi KL, Johnson KJ, Lu C et al: Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun 2014; 5: 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP et al: Germline disease-causing variants in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst 2015, e-pub ahead of print 27 August 2015 doi:10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed]
- Ren LP, Xian YS, Diao DM, Chen Y, Guo Q, Dang CX: Further evidence for the contribution of the BRCA1-interacting protein-terminal helicase 1 (BRIP1) gene in breast cancer susceptibility. Genet Mol Res 2013; 12: 5793–5801. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S et al: A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995; 268: 1749–1753. [DOI] [PubMed] [Google Scholar]
- Skorski T: Oncogenic tyrosine kinases and the DNA-damage response. Nat Rev Cancer 2002; 2: 351–360. [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Cicek MS, Larson NB et al: Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Sci Rep 2014; 4: 4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Hellebrand H, Wiek C et al: Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010; 42: 410–414. [DOI] [PubMed] [Google Scholar]
- Weissman SM, Weiss SM, Newlin AC: Genetic testing by cancer site: ovary. Cancer J 2012; 18: 320–327. [DOI] [PubMed] [Google Scholar]
- Sopik V, Akbari MR, Narod SA.: Genetic testing for RAD51C mutations: in the clinic and community. Clin Genet 2015; 88: 303–312. [DOI] [PubMed] [Google Scholar]
- Golmard L, Caux-Moncoutier V, Davy G et al: Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer 2013; 13: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers FS, Vreeswijk MP, van Asperen CJ, Devilee P: The impact of next generation sequencing on the analysis of breast cancer susceptibility: a role for extremely rare genetic variation? Clin Genet 2013; 84: 407–414. [DOI] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ramsay E et al: Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 2011; 43: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska MM, Samulak D, Smolarz B: An association between the -41657 C/T polymorphism of X-ray repair cross-complementing 2 (XRCC2) gene and ovarian cancer. Med Oncol 2014; 31: 300. [DOI] [PubMed] [Google Scholar]
- Park DJ, Lesueur F, Nguyen-Dumont T et al: Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet 2012; 90: 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Hewish M, Lord CJ, Ashworth A: Genomic instability and the selection of treatments for cancer. J Pathol 2010; 220: 281–289. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD et al: Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ et al: Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434: 917–921. [DOI] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ et al: Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006; 66: 8109–8115. [DOI] [PubMed] [Google Scholar]
- Evers B, Drost R, Schut E et al: Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res 2008; 14: 3916–3925. [DOI] [PubMed] [Google Scholar]
- Rottenberg S, Jaspers JE, Kersbergen A et al: High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA 2008; 105: 17079–17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT et al: Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010; 376: 245–251. [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE et al: Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376: 235–244. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C et al: Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014; 15: 852–861. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S et al: Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003; 72: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abugattas J, Llacuachaqui M, Allende YS et al: Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from Peru. Clin Genet 2014; 88: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Hwang UK, Kwak BS et al: Prevalence of BRCA1 and BRCA2 mutations in Korean breast cancer patients. J Korean Med Sci 2004; 19: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozek I, Ochman K, Debniak J et al: High frequency of BRCA1/2 germline mutations in consecutive ovarian cancer patients in Poland. Gynecol Oncol 2008; 108: 433–437. [DOI] [PubMed] [Google Scholar]
- De Leon Matsuda ML, Liede A, Kwan E et al: BRCA1 and BRCA2 mutations among breast cancer patients from the Philippines. Int J Cancer 2002; 98: 596–603. [DOI] [PubMed] [Google Scholar]
- Ginsburg OM, Dinh NV, To TV et al: Family history, BRCA mutations and breast cancer in Vietnamese women. Clin Genet 2011; 80: 89–92. [DOI] [PubMed] [Google Scholar]
- Hernández JE, Llacuachaqui M, Palacio GV et al: Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from medellin, Colombia. Hered Cancer Clin Pract 2014; 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi CE, van Ierland Y, van Asperen CJ et al: Prediction of BRCA1/2 mutation status in patients with ovarian cancer from a hospital-based cohort. Genet Med 2007; 9: 173–179. [DOI] [PubMed] [Google Scholar]
- Koumpis C, Dimitrakakis C, Antsaklis A et al: Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from Greece. Hered Cancer Clin Pract 2011; 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudico A, Redaniel MT, Mirasol-Lumague MR et al: Epidemiology and clinicopathology of breast cancer in metro Manila and Rizal Province, Philippines. Asian Pac J Cancer Prev 2009; 10: 167–172. [PubMed] [Google Scholar]
- Malander S, Ridderheim M, Masback A et al: One in 10 ovarian cancer patients carry germ line BRCA1 or BRCA2 mutations: results of a prospective study in Southern Sweden. Eur J Cancer 2004; 40: 422–428. [DOI] [PubMed] [Google Scholar]
- Menkiszak J, Gronwald J, Gorski B et al: Hereditary ovarian cancer in Poland. Int J Cancer 2003; 106: 942–945. [DOI] [PubMed] [Google Scholar]
- Risch HA, McLaughlin JR, Cole DE et al: Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 2001; 68: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch HA, McLaughlin JR, Cole DE et al: Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 2006; 98: 1694–1706. [DOI] [PubMed] [Google Scholar]
- Rodríguez AO, Llacuachaqui M, Pardo GG et al: BRCA1 and BRCA2 mutations among ovarian cancer patients from Colombia. Gynecol Oncol 2012; 124: 236–243. [DOI] [PubMed] [Google Scholar]
- Soegaard M, Kjaer SK, Cox M et al: BRCA1 and BRCA2 mutation prevalence and clinical characteristics of a population-based series of ovarian cancer cases from Denmark. Clin Cancer Res 2008; 14: 3761–3767.18559594 [Google Scholar]
- Balmaña J, Diez O, Rubio IT, Cardoso F.: BRCA in breast cancer: ESMO Clinical Practice Guidelines. Ann Oncol 2011; 22 (Suppl 6): vi31–vi34. [DOI] [PubMed] [Google Scholar]
- Evans DG, Howell A: Breast cancer risk-assessment models. Breast Cancer Res 2007; 9: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaiti B, Alarcon F, Andrieu N et al: A new scoring system in cancer genetics: application to criteria for BRCA1 and BRCA2 mutation screening. J Med Genet 2014; 51: 114–121. [DOI] [PubMed] [Google Scholar]
- Evans DG, Eccles DM, Rahman N et al: A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet 2004; 41: 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Lalloo F, Wallace A, Rahman N: Update on the Manchester Scoring System for BRCA1 and BRCA2 testing. J Med Genet 2005; 42: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaiti B, Alarcon F, Bonadona V et al: [A new scoring system for the diagnosis of BRCA1/2 associated breast-ovarian cancer predisposition]. Bull Cancer 2011; 98: 779–795. [DOI] [PubMed] [Google Scholar]
- Atchley DP, Albarracin CT, Lopez A et al: Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol 2008; 26: 4282–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Lalloo F, Cramer A et al: Addition of pathology and biomarker information significantly improves the performance of the Manchester scoring system for BRCA1 and BRCA2 testing. J Med Genet 2009; 46: 811–817. [DOI] [PubMed] [Google Scholar]
- Haffty BG, Yang Q, Reiss M et al: Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 2006; 24: 5652–5657. [DOI] [PubMed] [Google Scholar]
- Litwiniuk MM, Roznowski K, Filas V et al: Expression of estrogen receptor beta in the breast carcinoma of BRCA1 mutation carriers. BMC Cancer 2008; 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkevich V, Timms KM, Hennessy BT et al: Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012; 107: 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Wang ZC, Kim JY et al: Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012; 2: 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T, Manie E, Rieunier G et al: Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012; 72: 5454–5462. [DOI] [PubMed] [Google Scholar]
- Lancaster JM, Powell CB, Chen LM, Richardson DL: Society of Gynecologic Oncology statement on risk assessment for inherited gynecologic cancer predispositions. Gynecol Oncol 2015; 136: 3–7. [DOI] [PubMed] [Google Scholar]