Fig. 1.
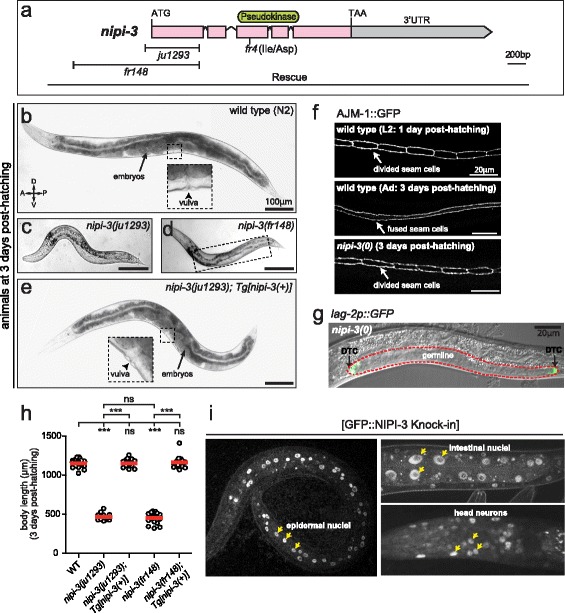
C. elegans Tribbles nipi-3 is required for larval development and viability. a The nipi-3 locus. Top, nipi-3 encodes a pseudokinase of the Tribbles family. Middle, nipi-3 deletions generated using CRISPR-Cas9 genome editing. Bottom, the extent of the nipi-3 genomic region used to rescue the deletion mutants. b–e Bright-field images of worms at 3 days post-hatching; wild type (b), nipi-3 null mutants (c,d) and a transgenic animal (Tg) expressing the wild-type nipi-3 genomic DNA in a nipi-3(0) background (e). f Fluorescence images of worms expressing AJM-1::GFP reporter in the epithelial cells to allow visualization of seam cells. g Overlaid differential interference contrast (DIC) and fluorescence image of a nipi-3(fr148) mutant at 3 days post-hatching expressing lag-2p::GFP reporter. This image corresponds to the boxed region in d. Two distal tip cells (DTC) are shown in green (arrow), and the germline of arrested nipi-3 null larva is denoted by a dotted red line. h Body length (μm) of worms at 3 days post-hatching. Each dot represents a single animal measured as shown; each red line represents the mean value. ***P < 0.001; ns, not significant (one-way ANOVA with Tukey’s post hoc tests). i Fluorescence images of endogenous NIPI-3 expression visualized in the GFP KI strain (fr152). Expression is observed in the nuclei (yellow arrows) of the epidermis (left panel), the intestine (upper right panel) and the head neurons (lower right panel)
