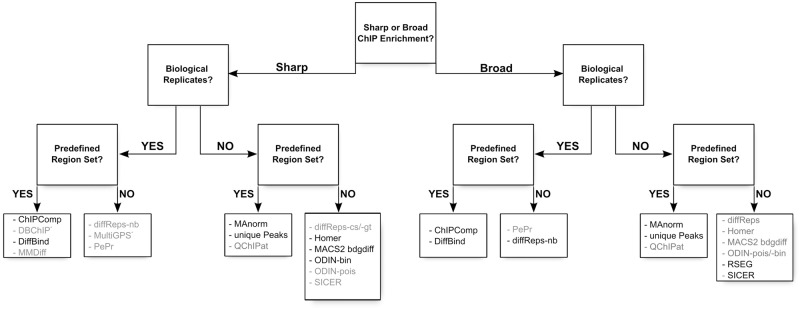
Figure 7.
Decision tree indicating the proper choice of tool depending on the data set: shape of the signal (sharp peaks or broad enrichments), presence of replicates and presence of an external set of regions of interest. We have indicated in dark the name of the tools that give good results using default settings, and in gray the tools that would require parameter tuning to achieve optimal results: some tools suffer from an excessive number of DR (PePr, ODIN-pois), an insufficient number of DR (QChIPat, MMDiff, DBChIP) or from an imprecise definition of the DR for sharp signal (SICER, diffReps-nb). *MultiGPS has been explicitly developed for transcription factor ChIP-seq.