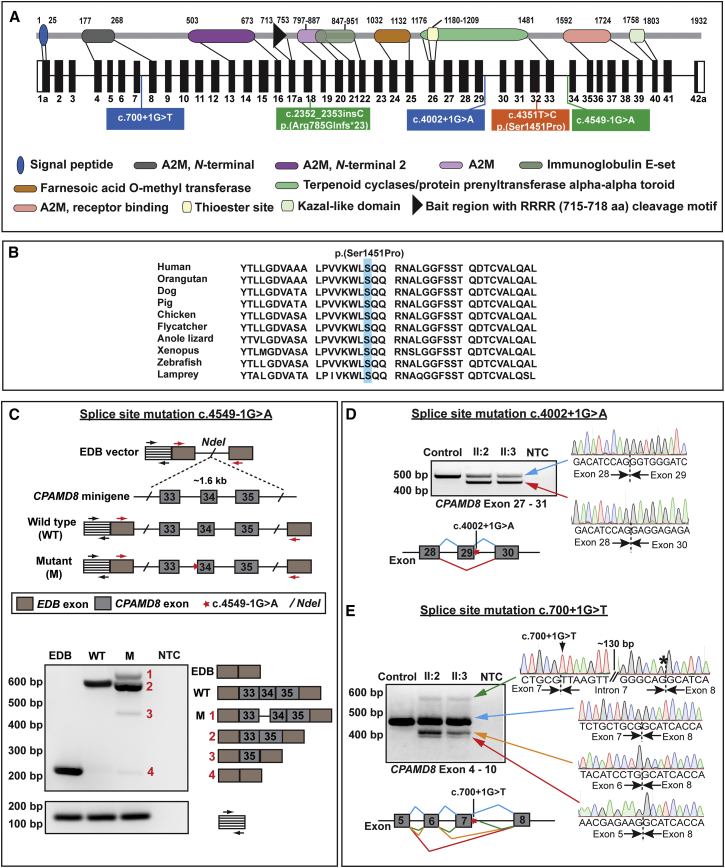
Figure 3.
Schematic of CPAMD8 Gene and Protein Structure and Consequence of CPAMD8 Mutations Identified in this Study
(A) Schematic of the genomic and protein structure of CPAMD8-1a. Positions of CPAMD8 mutations identified in this study are illustrated. The homozygous missense variant, p.Ser1404Pro, identified in family 1 is located in exon 32 (orange box). Compound heterozygous frameshift and splice-site variants, p.Arg785Glnfs∗23 and c.4549-1G>A, were identified in family 2 (green boxes). In family 3, compound heterozygous splice-site mutations, c.700+1G>T and c.4002+1G>A, were identified (blue boxes). Mutations are annotated in accordance with transcript CPAMD8-1a (GenBank: NM_015692.2). A2M, α2-macroglobulin.
(B) Multiple sequence alignment of CPAMD8 orthologs shows that the serine residue altered in family 1, p.Ser1451Pro, is highly conserved in different species.
(C) In vitro splice assay of CPAMD8 via a minigene system. Wild-type and mutant c.4549-1G>A fragments of CPAMD8 exons 33 to 35 with flanking intronic sequence were cloned into the EDB splice assay vector. The position of the mutation is highlighted with a red star. Primer binding sites to EDB exons are indicated by red arrows, whereas the control primers targeting the vector backbone sequences are depicted by black arrows. Transcripts generated by RT–PCR were separated by agarose gel electrophoresis and were directly sequenced. The wild-type construct generated a correctly spliced CPAMD8 exon 33-34-35 to the vector exons, whereas the mutant construct generated four aberrantly spliced products. The most efficiently spliced product (2) represents an aberrant splicing event that caused skipping of exon 34. Other aberrantly spliced products detected correspond to the inclusion of intron 33 (1) and the deletion of both exons 33 and 34 (3). The spliced product corresponding to the vector exons alone (4) was also produced by the mutant constructs at a very low level. Amplification with control primers (black arrows) demonstrates equal loading of the cDNA template. NTC, no template control.
(D and E) Agarose gel images showing RT-PCR products from whole-blood RNA of affected individuals II:2 and II:3 from family 3, in comparison to a control. Electropherograms and schematic show the transcripts generated by the c.4002+1G>A and c.700+1G>T mutations. Exons are indicated by filled bars and introns are depicted by lines. Splicing events are color coded according to the transcripts in the agarose gel images.
(D) Amplification of exons 27–31 encompassing the splice-donor site mutation c.4002+1G>A resulted in two transcripts in both affected siblings. Direct sequencing revealed wild-type product (blue arrow) and a smaller transcript of approximately 470 bp (red arrow), corresponding to exon 29 skipping, which resulted in an in-frame deletion, p.Gly1310_Glu1334del.
(E) Wild-type transcript is indicated by a blue arrow. Splice-donor mutation c.700+1G>T resulted in three aberrant transcripts. The top electropherogram (green arrow) shows activation of a cryptic splice-donor site (black asterisk) in intron 7 (142 bp downstream of exon 7), which is predicted to result in a prematurely truncated product, p.Gly234Valfs∗30. Two smaller transcripts (orange and red arrows) depict two additional abnormal splicing events, the skipping of exon 7 alone and skipping of exons 6 and 7, which result in frameshifts p.Asp216Alafs∗5 and p.Leu210Alafs∗5, respectively.