Figure 6.
Padi3 Gene Disruption in Mice
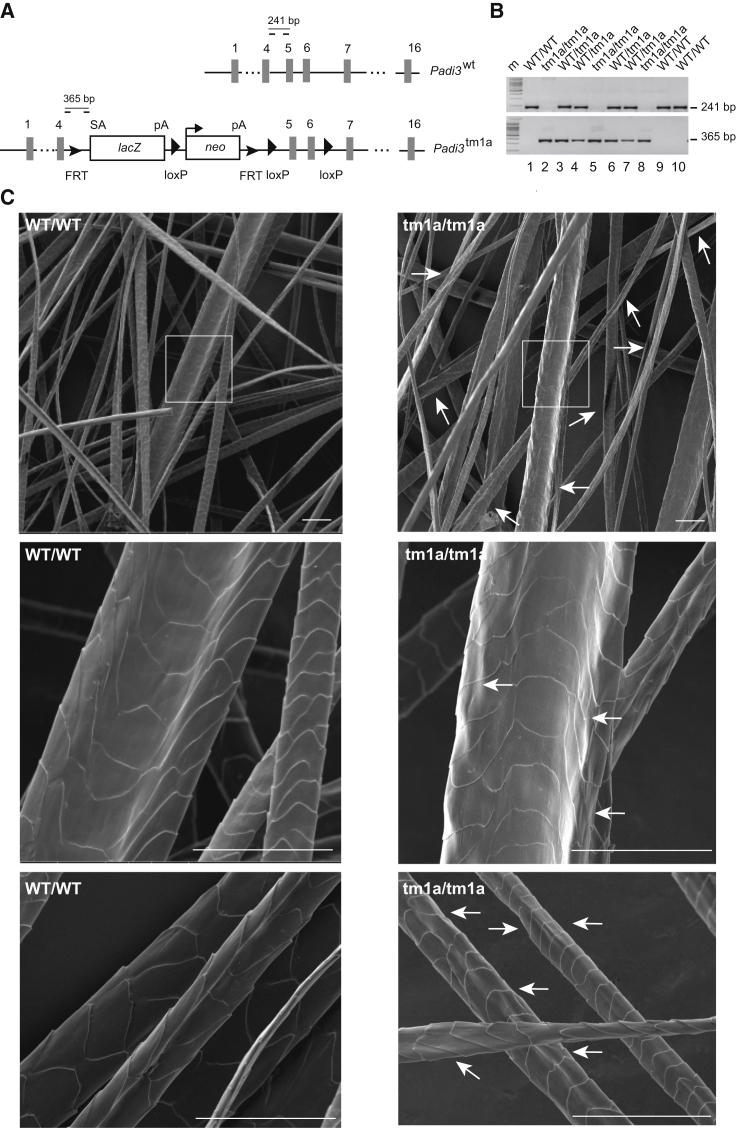
(A) Schematic representation of Padi3 and the targeting vector. The exons are shown by numbered gray rectangles. A cassette containing the beta-galactosidase (LacZ) and neomycin (neo) genes is inserted between exons 4 and 5. This results in the production of a truncated Padi3 protein corresponding to amino acids 1–136 fused to β-galactosidase. The position of the inserted loxP and flippase recognition target (FRT) sites are indicated, as well as the splicing acceptor site (SA) and the polyadenylation sites (pA) of LacZ and neo. The positions of the oligonucleotides (dashes) used for genotyping are shown, as well as the length of the amplified PCR products in wild-type (Padi3wt) and targeted (Padi3tm1a) alleles.
(B) PCR on genomic DNA from tail biopsies of ten mice from the same littermate (five females [1–5] and five males [6–10]) showing amplification of the 241 bp fragment for the Padi3wt allele and the 365 bp fragment for the Padi3tm1a allele.
(C) Representative pictures of SEM analysis of back hair coat from the three WT and three Padi3tm1a/tm1a mice (7 weeks old). Most hairs of the knockout mice are rough, with an irregular surface (white arrows). This is evident for thick hairs (top and middle micrographs) but was also observed for thin hairs (bottom). In some cases the hairs are twisted. The boxed areas are enlarged in the middle. At least 6 vibrissae and 200 hairs were analyzed per animal. Scale bars represent 50 μm.