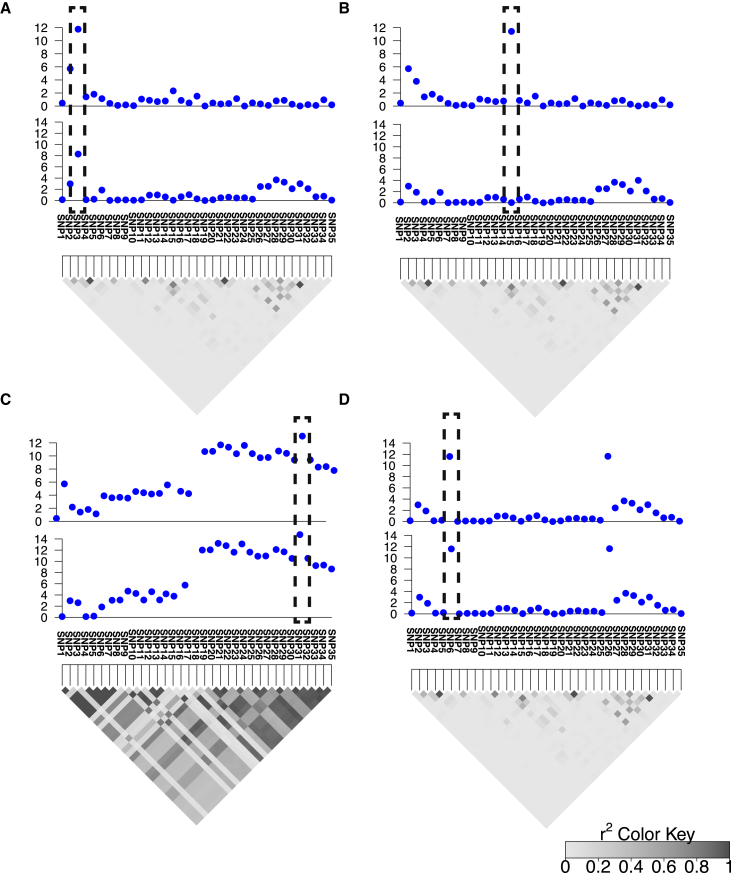
Figure 2.
Overview of eCAVIAR
Broadly, eCAVIAR aligns the causal variants in an eQTL study and GWAS. The x axis is the variant (SNP) location, and the y axis is the significance score (−log of p value) for each variant. The gray triangle indicates the LD structure, and every diamond in this triangle indicates the Pearson’s correlation. The darker the diamond, the higher the correlation; and the lighter the diamond, the lower the correlation between the variants.
(A) In the case where the causal variants are aligned, the colocalization posterior probability (CLPP) is high for the variant that is embedded in the dashed black rectangle.
(B) However, in the case where the causal variants are not aligned (the causal variants are not the same variants), the quantity of CLPP is low for the variant that is embedded in the dashed black rectangle.
(C) In this case, the LD is high, which implies that the uncertainty is high as a result of LD, and the CLPP value is low for the variant that is embedded in the dashed black rectangle.
(D) A case where a locus has two independent causal variants. If we consider that we have only one causal variant in a locus, then the CLPP of the causal variants is estimated to be 0.25. However, if we allow more than one causal variant in the locus, eCAVIAR estimates the CLPP to be 1.