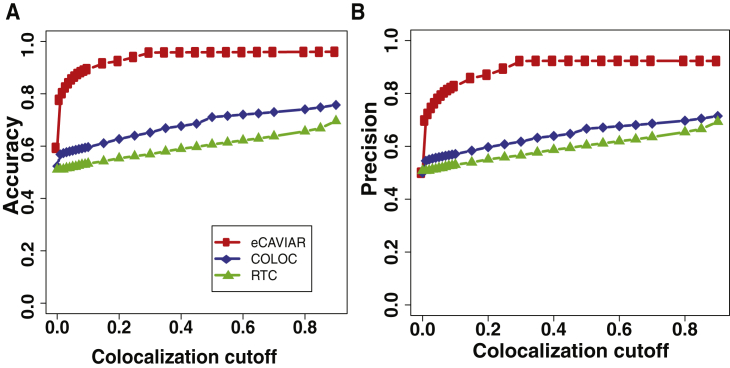
Figure 4.
eCAVIAR Is More Accurate Than Existing Methods for Regions with One Causal Variant
We compare the accuracy and precision of eCAVIAR with those of the two existing methods (RTC and COLOC). The x axis is the colocalization cutoff threshold. In these datasets, we implanted one causal variant, and we utilized simulated genotypes. We simulated the genotypes by using HAPGEN235 software. We used the European population from 1000 Genomes data33, 34 as the starting point to simulate the genotypes. The accuracy and precision of all three methods are shown in (A) and (B), respectively. We computed the TP (true-positive rate), TN (true-negative rate), FN (false-negative rate), and FP (false-positive rate) for the set of simulated datasets for which we generated the marginal statistics in a linear model. Accuracy = (TP + TN)/(TP + FP + FN + TN), and precision = TP/(TP + FP). We set the non-colocalization cutoff threshold to 0.001. We observed that eCAVIAR and COLOC had higher accuracy and precision than RTC.