Fig. 1.
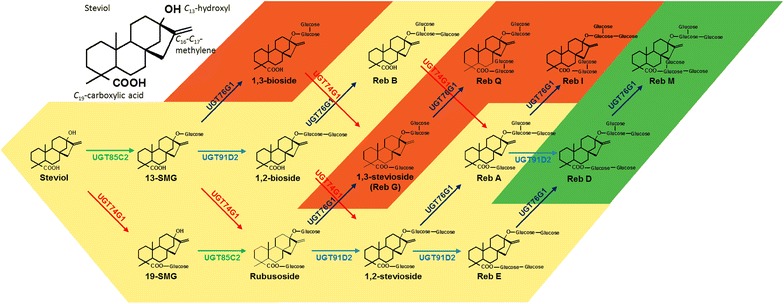
Steviol and the metabolic grid of glucosylation reactions resulting in formation of Reb D and Reb M. Top left the structure of steviol with emphasis on its functional groups. Reb D and Reb M are the two desired sweeteners (shown on green background). UGT91D2 has not been observed to glucosylate glucoside structures that harbor a glucose residue bound in a 1,3-glucosidic linkage. Formation of the 1,3-bond prior to formation of the 1,2-bond results in production of undesired side-products (shown on red background) [13, 15, 16]. UGT76G1 is known to glucosylate Steviol-13-O-monoglucoside (13-SMG), rubusoside, 1,2-stevioside and Reb D. In this study, 1,2-bioside, Reb G, Reb A and Reb E were identified as additional UGT76G1 substrates. UGT76G1 catalyzed glucosylation of Reb G and Reb A lead to the formation and structural elucidation of two new steviol glucosides Reb Q and Reb I
