Fig. 8.
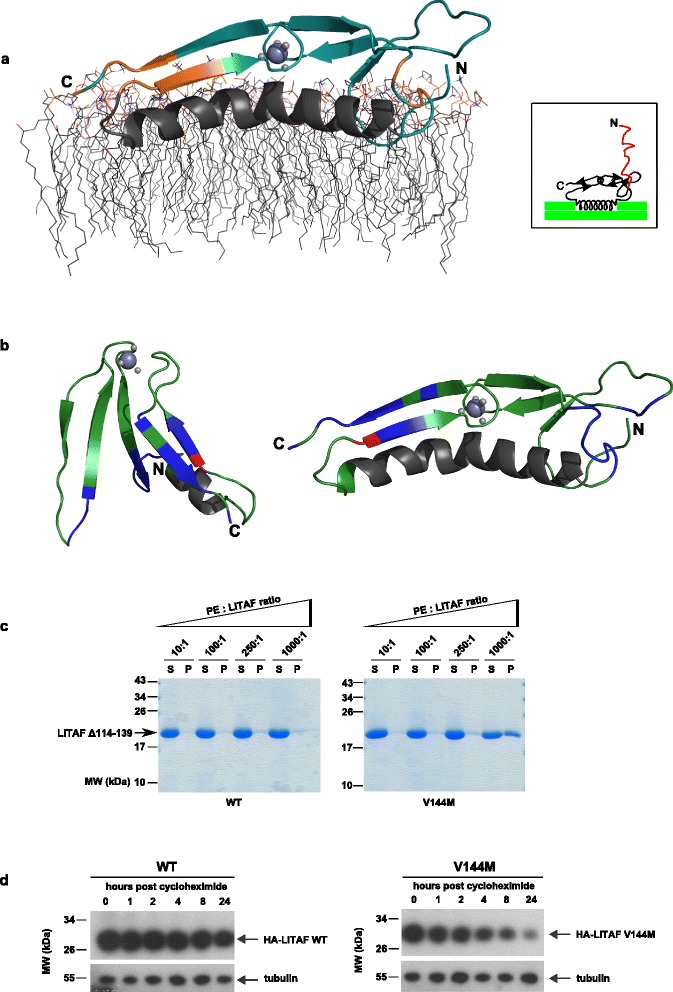
LITAF interacts with PE head groups and leads to protein instability in the V144M CMT1C-associated protein construct. a The position of residues displaying chemical shift perturbations greater than 1 standard deviation of the mean observed in the presence of PE head groups are coloured orange on the structural model of the LITAF domain. The illustration also shows the possible orientation of the LITAF domain as it inserts into a membrane containing phosphotidylethanolamine and phosphatidylcholine [85]. A cartoon diagram of the topology of the LITAF protein embedded in a lipid membrane bilayer (green), additionally showing the position of the flexible N-terminal proline-rich region (red), is shown in the inset. b The location of the residues displaying chemical shift perturbations greater than 1 standard deviation from the mean following the introduction of the V144M mutation are shown in blue on the structural models of the LITAF domain (LITAF Δ114–139 on the left and LITAF WT on the right). The position of Valine 144 is shown in red. c Equal amounts of LITAF Δ114–139 were incubated with increasing concentrations of PE for 2 hours at room temperature before centrifugation at 20,000 × g for 10 minutes to obtain soluble (S) and insoluble (P) fractions, respectively. The fractions were then separated by SDS-PAGE prior to staining with Coomassie Blue. In contrast to the WT construct (left panel), the V144M mutation rendered the protein unstable in the presence of higher concentrations of PE, leading to precipitation (right panel). d Retinal pigment epithelial cells stably expressing either HA-LITAF WT or HA-LITAF V144M were incubated with 300 μg/mL cycloheximide and incubated at 37 °C for the times indicated. Cellular lysates were prepared and the amount of HA-LITAF protein was determined by western blotting
