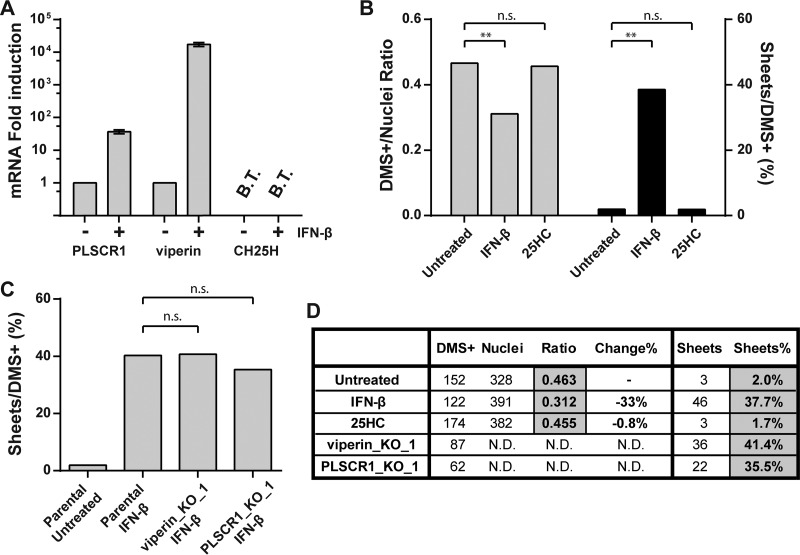
FIG 5 .
CH25H, PLSCR1, and viperin are not involved in the inhibition of EAV nsp2-3 DMV formation by IFN-β. (A) Relative induction of indicated ISG mRNAs in HuH-7 cells treated with IFN-β for 16 h compared to untreated control cells, using reverse transcriptase quantitative PCR (RT-qPCR) analysis. Indicated genes were amplified with gene-specific primers, error bars were based on three independent experiments. CH25H mRNA was below the threshold of detection (B.T.) in HuH-7 cells. The CH25H RT-qPCR was validated using cDNA of lipopolysaccharide-treated human dendritic cells. (B) Tetracycline-induced HuH-7/tetR/HA-nsp2-3GFP cells were treated with 500 U/ml IFN-β or 10 µM 25HC or untreated and analyzed by EM for the total number of cells positive for DMS (DMS+) as well as cell profiles positive for a nucleus (Nuclei). A section covering 300 to 500 cell profiles with a nucleus was analyzed for each sample. Ratios are calculated as the number of DMS+ cell profiles divided by the number of nucleus-positive cell profiles as well as the percentage of difference from the untreated control. The number of cell profiles positive for double-membrane sheets was also quantified, and the ratio of sheet-positive cell profiles compared to the total number DMS+ cell profiles was determined. (C) Tetracycline-induced CRISPR/Cas9 knockout cell lines of indicated genes were treated with 500 U/ml IFN-β and compared to the parental control cells. The analysis was similar to that for panel B, except that the DMS+/nucleus ratio was not determined. (D) A table of the quantifications shown in panels B and C. Statistical analyses were done using chi-square tests. **, P < 0.01. n.s., not significant; ND, not determined.