Figure 1.
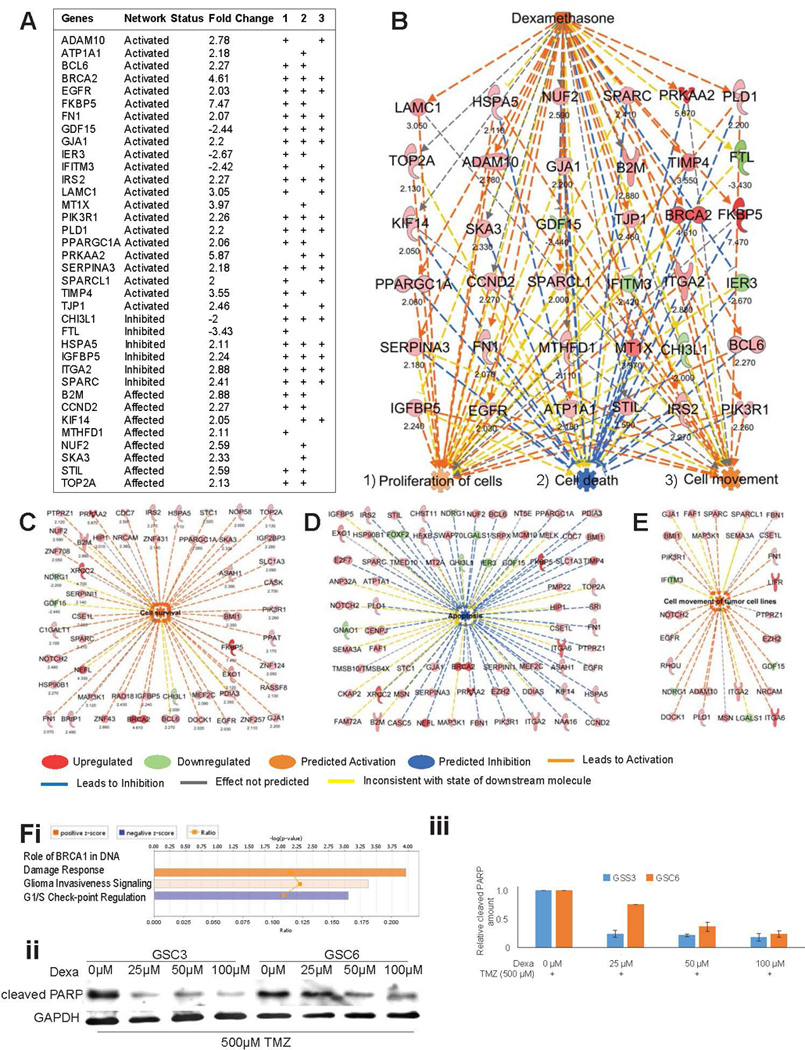
Apoptosis is inhibited and survival is activated upon dexamethasone exposure. (A) Fold-changes of gene expression in the dexamethasone network predicted its activation status. Their respective associations (+) with the cellular functions (1) proliferation, (2) cell death, and (3) cell movement (3) are shown. (B) This schematic representation shows the dexamethasone network, the genes involved in its activation, and its predicted impact on cellular functions. Numbers under the gene names indicate the fold-change in expression. (C–E) Individual genes contributing to the activated state of cell survival (C), the inhibited state of apoptosis (D), and the activated state of cell movement (E) in GSCs in response to dexamethasone are represented schematically. Numbers under the gene names indicate the fold-change in expression. Genes and cellular functions are labeled and color coded. A color index is shown at the bottom. (F) Network analysis for canonical pathway affected by dexamethasone shows “Role of BRCA1 in DNA Damage Response” with z-score 1.633 as top result (i). Scans of Western blots of whole-cell extracts of GSC3 and GSC6 treated with different concentrations of dexamethasone over three days under DNA damage stress of concomitant treatment with 500 µM Temozolomide and probed with an antibody against cleaved PARP as marker of apoptosis (GAPDH is loading control) (ii). Quantification of relative cleaved PARP amounts in western blots (shown in ii) normalized with GAPDH (iii).
