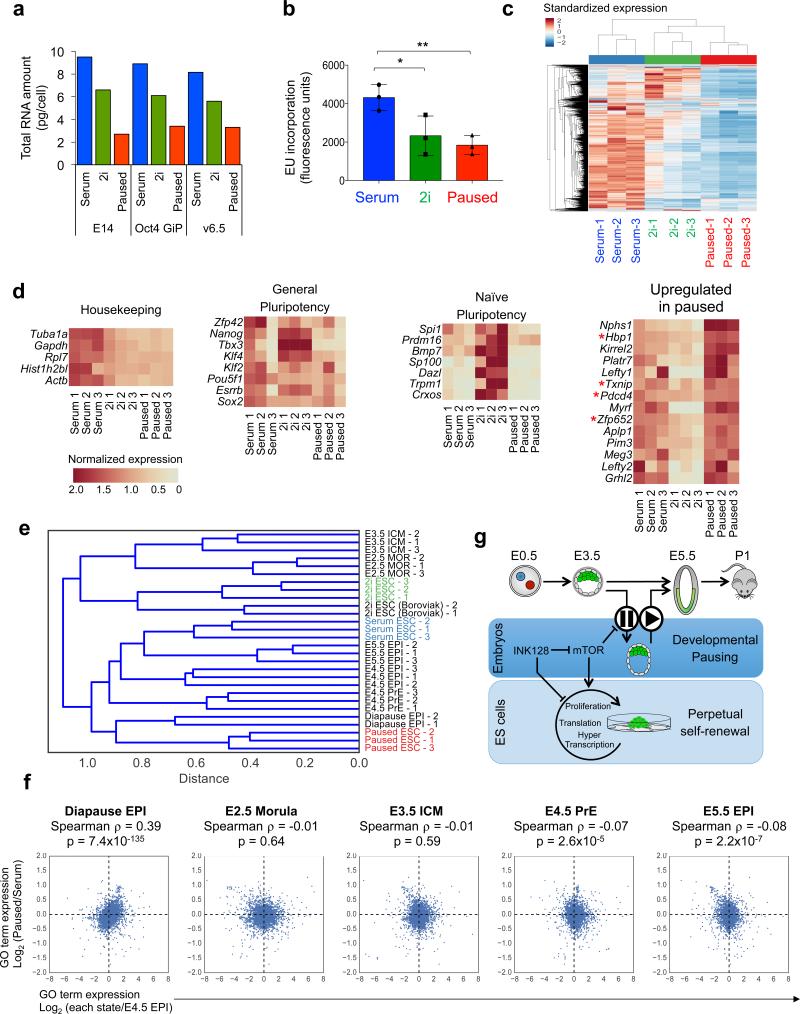
Figure 4.
Paused ES cells display global transcriptional suppression and mimic diapause. a, Total RNA amount per cell in three ES cell lines cultured in serum, 2i or pause conditions. The same cell lines were used for RNA-seq (c-e). b, Flow cytometry analysis of nascent transcription in the three states. Three technical replicates using E14 cells are shown. c, Hierarchical clustering showing standardized expression values for 5992 genes with robust expression in at least one ES cell state (see Methods). d, Heatmap representation of the indicated gene sets in serum, 2i and paused ES cells. Asterisks indicate repressor factors. e-f, Comparison of RNA-seq data from this study with distinct pluripotent cells in vivo20. e, Hierarchical clustering indicating that the transcriptome of paused ES cells is similar to the in vivo diapaused epiblast. f, Scatter plots of GO term expression. y-axis represents log2 fold change of GO term expression in paused vs. serum ES cells. x-axes represent log2 fold change of GO term expression in the indicated developmental stages vs. E4.5 epiblast. g, Model for the developmental pausing of blastocysts and ES cells by mTor inhibition.