Abstract
The timing of puberty onset varies greatly among individuals, and much of this variation is modulated by genetic factors. This study aimed to identify the kisspeptin receptor (KISS1R) gene variations and to investigate the associations between these variations and central precocious puberty (CPP). Korean girls with CPP (n = 194) and their healthy controls (n = 99) were included in this study. The entire coding region and the exon-intron boundaries (exon 1 through 5) of the KISS1R gene were directly sequenced. Seven polymorphisms were identified in the KISS1R gene. A missense change c.1091T>A, and an intron variant c.738+64G>T showed significantly higher allele frequencies in CPP patients than in controls (c.1091T>A: 30.7% vs. 22.2%, P = 0.031; c.738+64G>T: 45.6% vs. 35.9%, P = 0.023). The missense variant (c.1091T>A) was a nonsynonymous polymorphism that induces amino acid substitution of p.Leu364His. The haplotype CAGTGTC was detected more frequently in the CPP group (P = 0.042). The sequence variants of the KISS1R gene can be inducible factors in the development of CPP. The association between sequence variants and CPP should be validated by further evidence obtained from larger samples of children with CPP.
Keywords: KISS1R Gene, Precocious Puberty, Central, Polymorphism, Timing of Puberty
Graphical Abstract
INTRODUCTION
Human puberty is initiated by an increase in the amplitude and frequency of pulses of gonadotropin-releasing hormone (GnRH) after a quiescent period during childhood. The resurgence of pulsatile GnRH release from specialized hypothalamic neurons induces the secretion of gonadotropin by the pituitary gland, which leads to gonadal maturation and the attainment of reproductive capacity (1). Pubertal onset occurs across a wide range of ages in normal, healthy adolescents and is influenced by complex interactions among genetic, nutritional, environmental, and socioeconomic factors (2). The genetic factors influence pubertal timing in 50%–80% of the general population (3).
Precocious puberty is defined as the appearance of secondary sexual characteristics in girls younger than 8 years old and in boys younger than 9 years old. Central precocious puberty (CPP) is caused by the premature activation of the hypothalamic-pituitary-gonadal axis. CPP occurs more frequently in girls than in boys, and most of these cases are considered idiopathic (4,5,6). Numerous studies have suggested that genetic factors play important roles in CPP. Familial CPP can occur in up to 27.5% of cases and segregation analysis has suggested autosomal dominant transmission with incomplete sex-dependent penetrance (7). Recently, mutations in MKRN3, the gene encoding makorin RING-finger protein 3, have been shown to cause familial CPP (8). However, the genetic determinants of CPP are largely unknown.
Over the last decade, the kisspeptin/kisspeptin receptor (KISS1R) signaling complex has emerged as a fundamental regulator of puberty and reproductive function. KISS1R, one of the G protein-coupled receptors, is a 398 amino acid protein going through the membrane seven times and is activated by its ligand, kisspeptin. The receptor is expressed widely in the brain, pituitary gland, and placenta (9). In 2003, two research groups independently reported inactivating mutations of the KISS1R gene in patients with idiopathic hypogonadotropic hypogonadism (IHH) (10,11). These studies confirmed the critical role played by the kisspeptin/KISS1R system in hypothalamic-pituitary-gonadal axis activation. Navarro et al. (12) showed that binding to the G protein-coupled receptor in hypothalamic GnRH neurons enables kisspeptin to act as a powerful elicitor of GnRH-dependent luteinizing hormone (LH) secretion. In our previous study (13), we demonstrated that serum kisspeptin levels were significantly higher in girls with CPP than in controls. The KISS1 gene encoding kisspeptin and the KISS1R gene have recently been implicated in the pathogenesis of sexual precocity in humans. Activating mutations in KISS1 and KISS1R were detected as genetic causes of CPP (14,15). Accordingly, it is assumed that genetic variations in the KISS1R gene can lead to higher or lower risk of CPP. However, few mutations in the KISS1R gene that cause CPP have been identified in previous studies, with the exception of the study mentioned above.
The aims of our study were to evaluate the occurrence of genetic variations, including mutations and single-nucleotide polymorphisms (SNPs), of the KISS1R gene in 194 girls with CPP and to investigate the correlation between KISS1R gene variations and CPP in Korean girls.
MATERIALS AND METHODS
Subjects
Korean girls (n = 194) with idiopathic CPP were recruited from our outpatient clinic at Korea University Ansan Hospital, Korea between August 2012 and July 2013. The girls were diagnosed with idiopathic CPP if the following criteria were met: 1) objective breast budding appearing before the age of 8 years, 2) advanced bone age (BA) at least 1 year ahead of the chronological age (CA), 3) significantly higher peak LH values compared with the cutoff value of 5 mIU/mL according to a GnRH stimulation test conducted prior to the age of 9 years, and 4) a non-pathological brain magnetic resonance imaging (MRI). All the subjects showed normal brain MRI. BA was determined by a single observer according to the Greulich and Pyle method, and sexual maturity rating (SMR) was assessed by one pediatric endocrinologist according to the Tanner staging system.
The control group consisted of 99 healthy Korean girls who were recruited as volunteers on the basis of a freewill questionnaire concerning their breast development after the age of 9 years.
Genetic analysis
Genomic DNA was extracted from peripheral leukocytes of the study subjects using a DNA isolation kit (QIAamp DNA Blood Maxi prep kit; Qiagen, Valencia, CA, USA). The entire coding region and the exon-intron boundaries (exon 1 through 5) of the KISS1R gene (GenBank accession No. NM_032551.4) were amplified by polymerase chain reaction (PCR) with 12 pairs of specific primers (Table 1). The PCR conditions were as follows: initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C–65°C for 30 seconds, initial extension at 72°C for 30–60 seconds, and final extension at 72°C for 10 minutes. The PCR products were purified using a MultiScreen384-PCR Filter Plate (Milipore, Billerica, MA, USA). The purified products were then sequenced using a BigDye Terminator Cycle Sequencing Kit and an ABI 3730xl automated sequencer (Applied Biosystems, Foster City, CA, USA). The sequencing primers were the same as those used for the PCR amplification. Mutational analyses were performed using Phred, Phrap, Consed, and Polyphred 5.04 software (University of Washington, Seattle, WA, USA).
Table 1. Primers used in the analysis of the KISS1R gene.
| Exons | Primer sequences (5′ to 3′) | |
|---|---|---|
| Exon 1 | Forward | CAGGAAACAGCTATGACCGTGGGCAGGGGAGGGAGT |
| Reverse | TGTAAAACGACGGCCAGTACAGAGCCGGGTCCGAGA | |
| Exon 2 | Forward | CAGGAAACAGCTATGACCCTGGTCACTCGGACCAAGG |
| Reverse | TGTAAAACGACGGCCAGTCGTCCCCACGTACGATCC | |
| Exon 3 | Forward | CAGGAAACAGCTATGACCGTATGTGCCTGAGTGTTCG |
| Reverse | TGTAAAACGACGGCCAGTCTAGGCAGGCAGAGCTATT | |
| Exon 4 | Forward | CAGGAAACAGCTATGACCCAGGGTGGCTGGGTGAAC |
| Reverse | TGTAAAACGACGGCCAGTCTGGACCCCTTGGGCTGT | |
| Exon 5A | Forward | CAGGAAACAGCTATGACCGACAGCCCAAGGGGTCCA |
| Reverse | TGTAAAACGACGGCCAGTCGTTGTCCTCCCCCAGGA | |
| Exon 5B | Forward | CAGGAAACAGCTATGACCGCTGAACCCGCTGCTCTAC |
| Reverse | TGTAAAACGACGGCCAGTGTCTGGGTGACAGAATGAGACA | |
KISS1R = kisspeptin receptor.
Hormone assays
The GnRH stimulation test was conducted to evaluate pubertal status in all patients. Basal serum samples were obtained prior to GnRH (Relefact® LH-RH 0.1 mg; Aventis Pharma, Frankfurt, Germany) injection, and post-stimulation samples were acquired 30, 60, and 90 minutes after injection for measurements of LH and follicle-stimulating hormone (FSH) levels. The hormone levels were measured using a chemiluminescent microparticle immunoassay (Abbott Laboratories, Abbott Park, IL, USA).
Haplotype construction and statistical analysis
The allele frequencies were compared between the patient and control groups. Deviations from the Hardy-Weinberg equilibrium were tested by a comparison of observed and expected genotype frequencies. When the significant difference of allele frequencies between the patient and the control groups was found for each polymorphism, the clinical characteristics and results of the hormonal study were also compared between patients harboring a certain polymorphism and those lacking that polymorphism. Haploview 4.2 was used to calculate linkage disequilibrium (LD) (16) and standardized LD coefficient (D′) was plotted. The IBM SPSS 20.0 Network version program (IBM, Armonk, NY, USA) was used to perform statistical analyses, with P < 0.05 as the minimal level of statistical significance. Data were expressed as the means ± standard deviation (SD) or the standard deviation score (SDS). Fisher’s exact test was used for data analysis. The odds ratios for SNPs were calculated with respective 95% confidence intervals (CIs).
Ethics statement
This study was approved by the institutional review board of Korea University Ansan Hospital, Ansan, Korea (IRB number AS12131), and written informed consent was obtained from all subjects and their parents.
RESULTS
Clinical and hormonal characteristics of the study population
In the patient group, the mean age at diagnosis was 8.08 ± 0.77 years. The BA at diagnosis was 9.87 ± 0.98 years, and the mean difference from the CA was 1.79 ± 0.74 years. The mean height and body mass index (BMI) SDSs were 1.25 ± 0.88 and 0.55 ± 1.01, respectively. According to the results of the GnRH stimulation test, the basal and peak LH levels were 0.62 ± 0.70 IU/L and 13.80 ± 11.71 IU/L, respectively. The basal and peak FSH levels were 3.13 ± 3.54 IU/L and 15.12 ± 6.95 IU/L, respectively. The peak LH/FSH ratio was 1.02 ± 0.82. The mean estradiol level was 6.55 ± 5.34 pg/mL, and the mean SMR stage at diagnosis was 2.55 ± 0.82 for breast development.
In the control group, the mean CA and BA were 10.03 ± 0.58 and 10.55 ± 1.43 years. The mean BA advancement was 0.42 ± 1.13 years. The mean height and BMI SDSs were 0.65 ± 1.18 and 0.70 ± 1.11, respectively. Baseline characteristics and the results of hormone assays in the study population are shown in Table 2.
Table 2. Baseline characteristics and results of hormone assays in the study population.
| Auxological parameters | Patients (n = 194) | Control (n = 99) | P value |
|---|---|---|---|
| Age, yr | 8.08 ± 0.77 | 10.03 ± 0.58 | < 0.001 |
| Height, SDS | 1.25 ± 0.88 | 0.65 ± 1.18 | < 0.001 |
| Weight, SDS | 1.40 ± 1.25 | 1.17 ± 1.65 | 0.090 |
| BMI, SDS | 0.55 ± 1.01 | 0.70 ± 1.11 | 0.350 |
| MPH, SDS | −0.25 ± 0.75 | −0.08 ± 0.81 | 0.560 |
| BA, yr | 9.87 ± 0.98 | 10.55 ± 1.43 | 0.010 |
| BA advancement, yr | 1.79 ± 0.74 | 0.42 ± 1.13 | < 0.001 |
| SMR, stage | 2.55 ± 0.82 | ||
| Basal LH, IU/L | 0.62 ± 0.70 | ||
| Peak LH, IU/L | 13.80 ± 11.71 | ||
| Basal FSH, IU/L | 3.13 ± 3.54 | ||
| Peak FSH, IU/L | 15.12 ± 6.95 | ||
| Peak/basal LH | 50.91 ± 51.92 | ||
| Peak LH/FSH | 1.02 ± 0.82 | ||
| Estradiol, pg/mL | 6.55 ± 5.34 |
Data are shown as the mean ± standard deviation (SD).
SDS = standard deviation score, BMI = body mass index, MPH = mid parental height, BA = bone age, SMR = sexual maturity rating, LH = luteinizing hormone, FSH = follicle-stimulating hormone.
Identified SNPs in KISS1R gene analysis
Concerning the genetic analysis, seven SNPs were detected in the KISS1R gene (Table 3). One of these polymorphisms, c.*120C>G, was previously identified in Chinese girls with CPP (17) and was located in 3′UTR. Moreover, the c.24A>G was a synonymous polymorphism and was previously found in CPP girls (18,19). A missense variant c.1091T>A was previously reported in Korean girls with CPP (20) as well as in the study of another population (18). It was a nonsynonymous SNP that induces amino acid substitution of p.Leu364His. We assessed this variant using three in silico prediction algorithms, Polymorphism Phenotyping-2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/), Sorting Intolerant from Tolerance (SIFT, http://sift.jcvi.org/) and MutationTaster (http://www.mutationtaster.org/), and it was considered as a benign polymorphism in all three in silico analyses. The c.-123C>T was located in 5′UTR and the other three SNPs-c.245-34G>A, c.505+42T>G, and c.738+64G>T– were intron variants. Six of seven polymorphisms except one intron variant (c.505+42T>G) were identified in both the patient and control groups, and the results fitted the expectation by Hardy-Weinberg equilibrium. Although one intron variant (c.505+42T>G) was detected only in the CPP group, it did not induce an amino change and did not appear to affect the activity of the KISS1R. Clinical and laboratory characteristics in CPP patients with SNP are shown in Table 4. There were no specific clinical characteristics in the patient with each SNP.
Table 3. Polymorphisms of the KISS1R gene identified by sequencing (n = 293).
| No. | Polymorphism | Location | mRNA position | dbSNP ID | MAF in samples | Note |
|---|---|---|---|---|---|---|
| 1 | c.-123C>T | Exon 1 | 39 | rs3810423 | 0.061 | 5′UTR |
| 2 | c.24A>G | Exon 1 | 185 | rs10407968 | 0.137 | Synonymous |
| 3 | c.245-34G>A | Intron 1 | - | rs117902808 | 0.177 | - |
| 4 | c.505+42T>G | Intron 3 | - | rs543723928 | 0.002 | - |
| 5 | c.738+64G>T | Intron 4 | - | rs350131 | 0.423 | - |
| 6 | c.1091T>A | Exon 5 | 1,252 | rs350132 | 0.278 | p.Leu364His |
| 7 | c.*120C>G | Exon 5 | 1,478 | rs3746146 | 0.063 | 3′UTR |
The positions of the polymorphisms are defined according to NM_032551.4.
KISS1R = kisspeptin receptor, MAF = minor allele frequency, UTR = untranslated region.
Table 4. Clinical and laboratory characteristics in CPP patients with each polymorphism.
| No. | SNP | Time of diagnosis, yr | SMR | LH, mIU/mL | FSH, mIU/mL | Peak LH/FSH ratio | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CA | BA | Basal | Peak | Basal | Peak | |||||
| 1 | c.-123C>T | - | 7.8 | 9.2 | 2.5 | 0.5 | 11.8 | 2.6 | 16.8 | 0.77 |
| 2 | c.24A/G | Synonymous | 7.9 | 9.6 | 2.4 | 0.4 | 10.8 | 2.8 | 15.4 | 0.77 |
| 3 | c.245-34G>A | - | 8.1 | 9.8 | 2.8 | 0.8 | 15.4 | 3.7 | 15.9 | 1.07 |
| 4 | c.505+42T>G | - | 8.1 | 9.8 | 2.7 | 0.6 | 13.9 | 3.0 | 15.4 | 1.01 |
| 5 | c.738+64G>T | - | 8.2 | 9.8 | 2.5 | 0.7 | 14.5 | 3.0 | 15.3 | 1.07 |
| 6 | c.1091T>A | p.Leu364His | 7.9 | 9.5 | 2.4 | 0.3 | 11.9 | 2.3 | 13.9 | 0.94 |
| 7 | c.*120C>G | - | 8.1 | 9.7 | 2.5 | 0.6 | 13.7 | 3.0 | 15.5 | 0.99 |
The positions of the polymorphisms are defined according to NM_032551.4. Data are shown as the mean value.
CPP = central precocious puberty, SNP = single-nucleotide polymorphism, LH = luteinizing hormone, FSH = follicle-stimulating hormone, CA = chronological age, BA = bone age, SMR = sexual maturity rating.
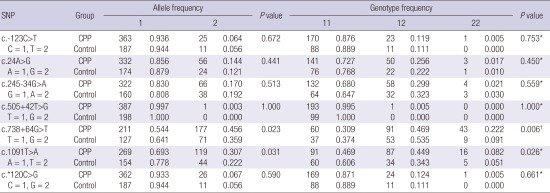
Allele counts and frequencies in the two groups are shown in Table 5. With Fisher’s exact test, the associations between the polymorphisms and the two phenotype groups were investigated. Among the polymorphisms, two SNPs, c.738+64G>T and c.1091T>A, were detected significantly more frequently allele frequencies in the CPP group (c.738+64G>T, P = 0.023; c.1091T>A, P = 0.031). Using the T allele of the intron variant c.738+64G>T as the reference, the odds ratio of the G allele for CPP was 1.501 (95% CI, 1.055–2.135). Also, using the A allele of the missense variant c.1091T>A as the reference, the odds ratio of the T allele for CPP was 1.548 (95% CI, 1.039–2.306). No differences were found between the two groups in the frequencies of the other five polymorphisms.
Table 5. Allele and genotype frequencies of the KISS1R polymorphisms.
| SNP | Group | Allele frequency | P value | Genotype frequency | P value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 11 | 12 | 22 | |||||||||
| c.-123C>T | CPP | 363 | 0.936 | 25 | 0.064 | 0.672 | 170 | 0.876 | 23 | 0.119 | 1 | 0.005 | 0.753* |
| C=1, T=2 | Control | 187 | 0.944 | 11 | 0.056 | 88 | 0.889 | 11 | 0.111 | 0 | 0.000 | ||
| c.24A>G | CPP | 332 | 0.856 | 56 | 0.144 | 0.441 | 141 | 0.727 | 50 | 0.256 | 3 | 0.017 | 0.450* |
| A=1, G=2 | Control | 174 | 0.879 | 24 | 0.121 | 76 | 0.768 | 22 | 0.222 | 1 | 0.010 | ||
| c.245-34G>A | CPP | 322 | 0.83 | 66 | 0.170 | 0.513 | 132 | 0.680 | 58 | 0.299 | 4 | 0.021 | 0.559* |
| G=1, A=2 | Control | 160 | 0.808 | 38 | 0.192 | 64 | 0.647 | 32 | 0.323 | 3 | 0.030 | ||
| c.505+42T>G | CPP | 387 | 0.997 | 1 | 0.003 | 1.000 | 193 | 0.995 | 1 | 0.005 | 0 | 0.000 | 1.000* |
| T=1, G=2 | Control | 198 | 1.000 | 0 | 0.000 | 99 | 1.000 | 0 | 0.000 | 0 | 0.000 | ||
| c.738+64G>T | CPP | 211 | 0.544 | 177 | 0.456 | 0.023 | 60 | 0.309 | 91 | 0.469 | 43 | 0.222 | 0.006† |
| T=1, G=2 | Control | 127 | 0.641 | 71 | 0.359 | 37 | 0.374 | 53 | 0.535 | 9 | 0.091 | ||
| c.1091T>A | CPP | 269 | 0.693 | 119 | 0.307 | 0.031 | 91 | 0.469 | 87 | 0.449 | 16 | 0.082 | 0.026* |
| A=1, T=2 | Control | 154 | 0.778 | 44 | 0.222 | 60 | 0.606 | 34 | 0.343 | 5 | 0.051 | ||
| c.*120C>G | CPP | 362 | 0.933 | 26 | 0.067 | 0.590 | 169 | 0.871 | 24 | 0.124 | 1 | 0.005 | 0.661* |
| C=1, G=2 | Control | 187 | 0.944 | 11 | 0.056 | 88 | 0.889 | 11 | 0.111 | 0 | 0.000 | ||
KISS1R = kisspeptin receptor, SNP = single-nucleotide polymorphism, CPP = central precocious puberty.
*P value for dominant model; †P value for recessive model.
Clinical significance of c.1091T>A (p.Leu364His) and c.738+64G>T in CPP patients
Based on the finding that two SNPs, c.1091T>A and c.738+64G>T, were detected more frequently in the patient group, we compared the clinical characteristics and hormone values between the two subgroups of these polymorphisms (c.1091T>A: the subgroup with AA and the subgroup with A/T or T/T, c.738+64G>T: the subgroup with TT or T/G and the subgroup with G/G). The subgroups of each polymorphism were classified according to the results of genotype frequencies (dominant model for c.1091T>A, and recessive model for c.738+64G>T). Basal and peak hormone levels, the LH/FSH ratio, and the auxological parameters at diagnosis—including CA, height, weight, BMI, mid parental height (MPH), SMR, and BA advancement—did not differ between the two subgroups of each polymorphisms in CPP patients. Data was not shown.
LD and haplotype analysis
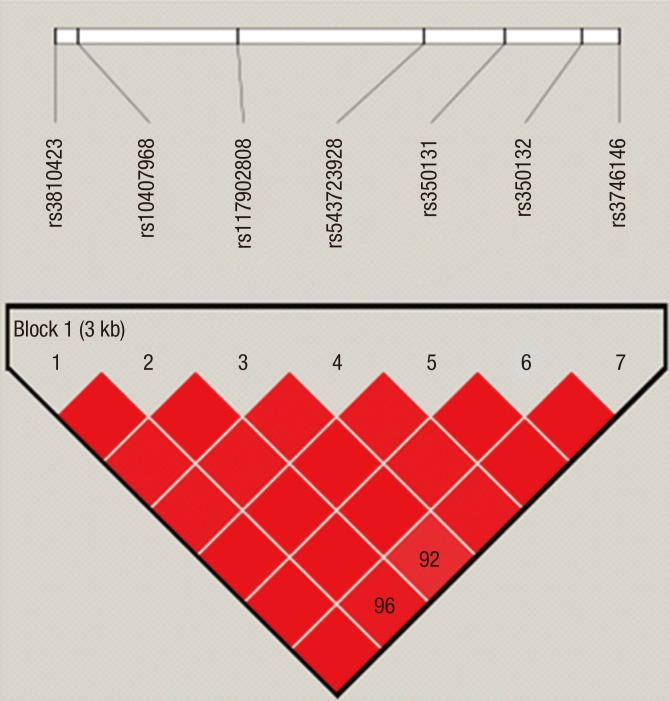
LD was computed between every two SNPs to further analyze the haplotype structure. Fig. 1 shows the LD plot constructed using the 7 SNPs. The magnitude of LD between each SNP was extremely high, with pair-wise D′ values that were ≥ 0.92. Five haplotypes were constructed based on the LD plot and the results are shown in Table 6. Haplotypes with frequency less than 1% are not listed. The haplotype CAGTGTC was detected statistically significantly more frequently in the CPP group with a moderate P value (0.042). The other four haplotypes showed no differences in frequencies between the patients and control groups.
Fig. 1.
LD plot for 7 SNPs in the KISS1R gene. The D′ values that correspond to each SNP pair are expressed as a percentage and shown within the respective square. Squares without numbers represent D′ values of 1.0, indicative of complete LD. The 7 SNPs constitute one haplotype block (Block 1).
LD = linkage disequilibrium, KISS1R = kisspeptin receptor, D′ = linkage disequilibrium coefficient, SNP = single-nucleotide polymorphism.
Table 6. Results of the haplotype analysis.
| Haplotypes* | CPP (n = 194) | Control (n = 99) | P value | ||
|---|---|---|---|---|---|
| Counts | Frequencies | Counts | Frequencies | ||
| CAATTAC | 64 | 0.165 | 38 | 0.323 | 0.415 |
| CAGTGTC | 117 | 0.302 | 44 | 0.222 | 0.042 |
| CAGTTAC | 147 | 0.379 | 89 | 0.450 | 0.099 |
| CGGTGAC | 30 | 0.077 | 13 | 0.066 | 0.609 |
| TGGTGAG | 25 | 0.064 | 11 | 0.056 | 0.672 |
CPP = central precocious puberty, SNP = single-nucleotide polymorphism.
*Haplotypes are designated according to the sequence of each SNP position as follows: rs3810423, rs10407968, rs117902808, rs543723928, rs350131, rs350132, rs3746146.
DISCUSSION
In this study, we evaluated a population of 194 girls with CPP for KISS1R gene mutations and polymorphisms, and we identified seven SNPs. Six polymorphisms except the intron variant c.505+42T>G were detected in both the CPP and control groups. Two polymorphisms, c.1091T>A and c.738+64G>T, were found to be statistically correlated to CPP. The missense variant (c.1091T>A) was a nonsynonymous polymorphism that induces amino acid substitution of p.Leu364His. The haplotype CAGTGTC was also significantly associated with CPP patients.
The pulsatile GnRH secretion mediated through the kisspeptin/KISS1R system appears to be a key event at the onset and during progression of puberty (11,21). The timing of puberty varies greatly among individuals and races, and much of this variation is due to genetic factors. The genetic modulation of puberty is assumed to arise from the additive effect of multiple genes (22). Many researchers have tried to ascertain a molecular mechanism of kisspeptin/KISS1R system associated with development of CPP. Silveira et al. (15) by investigating 83 children with CPP reported two novel KISS1 missense mutations in 3 unrelated children with CPP. Moreover, Ko et al. (23) identified one polymorphism (p.P110T) relevant to CPP in KISS1 gene in 101 Koreans girls with CPP and the polymorphism was suggested to exert a protective effect. In our previous study, we demonstrated that three KISS1 polymorphisms were statistically correlated with CPP, but could not find any clinical importance of these SNPs (24). Recently, it was found that the KISS1R gene is relevant to CPP pathogenesis (14). An autosomal dominant missense mutation, Arg386Pro, in KISS1R, leading to prolonged activation of intracellular pathways in response to kisspeptin, has been suggested to cause CPP in an adopted girl. Since then, no other CPP cases with activating KISS1R mutations have been reported. On the other hand, a few KISS1R polymorphisms have been identified in CPP patients. Some researchers have found A/G synonymous SNP (c.24A>G) in girls with CPP, but it did not seem to have a pathogenic role in patients (18,19). The polymorphism c.*120C>G has been reported in Chinese girls with CPP and the SNP was not statistically correlated with CPP (17). In the same study, the missense mutation p.P196H in the KISS1R was also identified in one patient, although functional study was not performed (17). Moreover, a missense variant c.1091T>A has been identified in studies of Korean girls with CPP and other ethnic group (18,20). However, the researchers were unable to determine any disease associations with the SNP.
In the present study, the seven polymorphisms were detected via the sequencing of the KISS1R gene. According to the calculated statistical results, a missense variant c.1091T>A exhibited a statistical correlation with the phenotype (P = 0.031). This polymorphism induced amino acid substitution of p.Leu364His. This SNP was previously reported in a study of Korean CPP patients in 2011 (20) and the allele frequency in the study was 30.3%. This result was slightly different to that in our study, in which the allele frequency was calculated as 27.8%. Since both study subjects had same ethnic background, the reason of this discrepancy in the allele frequency is unclear. One possible reason is a different sample size. In the previous study, the results showed a similar allele frequency in patients (30.3%) and controls (30.8%) (20). However, in our study, the allele frequencies were significantly higher in the patients (30.7%) than in the controls (22.2%), although the polymorphism c.1091T>A was identified in both the patients and the controls. Our finding suggests that the missense variant c.1091T>A might affect the pathogenesis of CPP. In order to evaluate the clinical significance of p.Leu364His, we compared the clinical characteristics and hormone values between the subgroups with or without this SNP in the patient group. However, no significant differences were observed between the two subgroups.
The intron variant c.738+64G>T also exhibited a statistical correlation with CPP patients (P = 0.023). It was located in the fourth intron with G to T substitution at nucleotide +64 and did not result in any amino acid change. This SNP was not introduced in previous studies concerning CPP. Although introns are generally known as non-protein-coding sections in genes, it can produce genetic and phenotypical variety by regulating or facilitating the transposition of exons (25). Therefore, it can be hypothesized that the polymorphism c.738+64G>T may affect the gene expression of KISS1R relevant to the pathogenesis of CPP in Korean girls. In the comparison of clinical parameters and hormonal results, no significant differences were observed between the subgroups with or without this SNP in the patient group. More evidence will be necessary to confirm the association between intron variant in the KISS1R and CPP.
In the current study, we have also identified a synonymous SNP (c.24A>G) residing in exon 1 of the KISS1R gene. This SNP was not correlated with CPP in our study. Since this variant induced a synonymous change, it did not appear to affect the activity of the KISS1R. This polymorphism was previously identified by others in a population of male patients with IHH, although no association with this disorder was found (26,27). It was also detected in CPP patients in previous studies, but it did not seem to have a pathogenic effect on CPP (18,19). These findings were in accord with our results. However, this polymorphism has not been recorded in a large population of Chinese girls (24 girls randomly selected from 272 girls with CPP) (17). The discordance in data between Korean and Chinese populations is partly due to different scale of typed samples. Also, the genetic background appeared to differ significantly, even though Koreans and Chinese are both Asian.
We identified one LD block that consisted of seven SNPs. The value of LD (D′) between each SNP was very high and five haplotype were constructed using the LD block. The haplotype CAGTGTC was detected more frequently in the CPP group (30.2%) than in the control group (22.2%) (P = 0.042). Our findings suggest that the haplotype CAGTGTC contributes to pathogenesis of CPP, although the P value of 0.042 was not sufficiently significant.
In conclusion, we have attempted to identify sequence variations of the KISS1R gene associated with CPP in Korean girls, and have noted some possibilities that the sequence variants within the KISS1R gene might be inducible factors in the pathogenesis of CPP. A possible limitation of the present study was that the sample scale was relatively small, and that P values were not very significant. Therefore, larger scale studies are necessary in order to clarify the association between the sequence variations of KISS1R and CPP.
Footnotes
Funding: This work was supported by a Korea University grant (2012) and research funding from the Korean Society of Pediatric Endocrinology (2012-02).
DISCLOSURE: The authors have no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTION: Research conception and design: Oh YJ, Rhie YJ, Lee KH. Performing the experiments: Oh YJ, Rhie YJ, Nam HK. Data acquisition: Nam HK, Rhie YJ, Kim HR. Data analysis and interpretation: Oh YJ, Rhie YJ, Lee KH. Statistical analysis: Oh YJ. Drafting of the manuscript: Oh YJ, Rhie YJ. Critical revision of the manuscript: Lee KH. Receiving grant: Rhie YJ, Lee KH. Approval of final manuscript: all authors.
References
- 1.Tena-Sempere M. The roles of kisspeptins and G protein-coupled receptor-54 in pubertal development. Curr Opin Pediatr. 2006;18:442–447. doi: 10.1097/01.mop.0000236396.79580.cc. [DOI] [PubMed] [Google Scholar]
- 2.Delemarre-van de Waal HA. Secular trend of timing of puberty. Endocr Dev. 2005;8:1–14. doi: 10.1159/000084082. [DOI] [PubMed] [Google Scholar]
- 3.Gajdos ZK, Henderson KD, Hirschhorn JN, Palmert MR. Genetic determinants of pubertal timing in the general population. Mol Cell Endocrinol. 2010;324:21–29. doi: 10.1016/j.mce.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teilmann G, Pedersen CB, Jensen TK, Skakkebaek NE, Juul A. Prevalence and incidence of precocious pubertal development in Denmark: an epidemiologic study based on national registries. Pediatrics. 2005;116:1323–1328. doi: 10.1542/peds.2005-0012. [DOI] [PubMed] [Google Scholar]
- 5.Brito VN, Latronico AC, Arnhold IJ, Mendonça BB. Update on the etiology, diagnosis and therapeutic management of sexual precocity. Arq Bras Endocrinol Metabol. 2008;52:18–31. doi: 10.1590/s0004-27302008000100005. [DOI] [PubMed] [Google Scholar]
- 6.Kakarla N, Bradshaw KD. Disorders of pubertal development: precocious puberty. Semin Reprod Med. 2003;21:339–351. doi: 10.1055/s-2004-815590. [DOI] [PubMed] [Google Scholar]
- 7.de Vries L, Kauschansky A, Shohat M, Phillip M. Familial central precocious puberty suggests autosomal dominant inheritance. J Clin Endocrinol Metab. 2004;89:1794–1800. doi: 10.1210/jc.2003-030361. [DOI] [PubMed] [Google Scholar]
- 8.Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368:2467–2475. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, et al. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 10.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 12.Navarro VM, Castellano JM, García-Galiano D, Tena-Sempere M. Neuroendocrine factors in the initiation of puberty: the emergent role of kisspeptin. Rev Endocr Metab Disord. 2007;8:11–20. doi: 10.1007/s11154-007-9028-2. [DOI] [PubMed] [Google Scholar]
- 13.Rhie YJ, Lee KH, Eun SH, Choi BM, Chae HW, Kwon AR, Lee WJ, Kim JH, Kim HS. Serum kisspeptin levels in Korean girls with central precocious puberty. J Korean Med Sci. 2011;26:927–931. doi: 10.3346/jkms.2011.26.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silveira LG, Noel SD, Silveira-Neto AP, Abreu AP, Brito VN, Santos MG, Bianco SD, Kuohung W, Xu S, Gryngarten M, et al. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab. 2010;95:2276–2280. doi: 10.1210/jc.2009-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Luan X, Yu H, Wei X, Zhou Y, Wang W, Li P, Gan X, Wei D, Xiao J. GPR54 polymorphisms in Chinese girls with central precocious puberty. Neuroendocrinology. 2007;86:77–83. doi: 10.1159/000107511. [DOI] [PubMed] [Google Scholar]
- 18.Krstevska-Konstantinova M, Jovanovska J, Tasic VB, Montenegro LR, Beneduzzi D, Silveira LF, Gucev ZS. Mutational analysis of KISS1 and KISS1R in idiopathic central precocious puberty. J Pediatr Endocrinol Metab. 2014;27:199–201. doi: 10.1515/jpem-2013-0080. [DOI] [PubMed] [Google Scholar]
- 19.Leka-Emiri S, Louizou E, Kambouris M, Chrousos G, De Roux N, Kanaka-Gantenbein C. Absence of GPR54 and TACR3 mutations in sporadic cases of idiopathic central precocious puberty. Horm Res Paediatr. 2014;81:177–181. doi: 10.1159/000356913. [DOI] [PubMed] [Google Scholar]
- 20.Ko JM, Lee HS, Lee HS, Hwang JS. Genetic variations of GNRH1, GNRHR and GPR54 genes in Korean girls with central precocious puberty. J Korean Soc Pediatr Endocrinol. 2011;16:38–45. [Google Scholar]
- 21.Semple RK, Achermann JC, Ellery J, Farooqi IS, Karet FE, Stanhope RG, O’Rahilly S, Aparicio SA. Two novel missense mutations in g protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2005;90:1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- 22.Palmert MR, Boepple PA. Variation in the timing of puberty: clinical spectrum and genetic investigation. J Clin Endocrinol Metab. 2001;86:2364–2368. doi: 10.1210/jcem.86.6.7603. [DOI] [PubMed] [Google Scholar]
- 23.Ko JM, Lee HS, Hwang JS. KISS1 gene analysis in Korean girls with central precocious puberty: a polymorphism, p.P110T, suggested to exert a protective effect. Endocr J. 2010;57:701–709. doi: 10.1507/endocrj.k10e-073. [DOI] [PubMed] [Google Scholar]
- 24.Rhie YJ, Lee KH, Ko JM, Lee WJ, Kim JH, Kim HS. KISS1 gene polymorphisms in Korean girls with central precocious puberty. J Korean Med Sci. 2014;29:1120–1125. doi: 10.3346/jkms.2014.29.8.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapp G. Exons, introns, and talking genes: the science behind the human genome project (book) Libr J. 1991;116:114–118. [Google Scholar]
- 26.Tenenbaum-Rakover Y, Commenges-Ducos M, Iovane A, Aumas C, Admoni O, de Roux N. Neuroendocrine phenotype analysis in five patients with isolated hypogonadotropic hypogonadism due to a L102P inactivating mutation of GPR54. J Clin Endocrinol Metab. 2007;92:1137–1144. doi: 10.1210/jc.2006-2147. [DOI] [PubMed] [Google Scholar]
- 27.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]