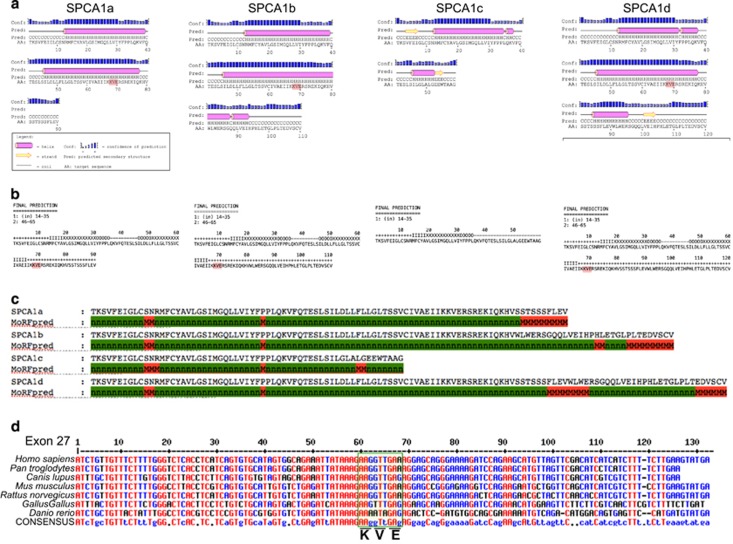
Figure 4.
Bioinformatics analysis of SPCA1 isoforms C-terminal region. (a) Secondary structure propensities in the proteins' regions encoded by terminal exons were predicted with the program PSIPRED.92 Highlighted in pink is the corresponding sequences in the KXD/E motif. Predictions indicate that this motif is located in a helical extension spanning outside transmembrane helix M10 (see also b). Interestingly this helix is predicted to have different length in the three isoforms where it is present (a, b and d). (b) Transmembrane topology prediction was performed using the algorithms MEMSAT3 (ref. 93) and TMHMM2 (http://www.cbs.dtu.dk/services/TMHMM) yielding very similar results. In the isoforms SPCA1a, SPCA1b and SPCA1d, two transmembrane helices are predicted by both algorithms, the first (M9) with cytosolic-lumenal orientation and the second (M10) with opposite orientation. In the SPCA1c isoform a single cytosolic-lumenal transmembrane helix (M9) is predicted. Shown are results from MEMSAT3 where ‘+' stands for cytosolic residue, ‘−' for lumenal residue and I, X and O for transmembrane residues (I close to cytosolic, X central and O close to lumenal). (c) The MoRFPred algorithm94 was used to predict, in the different isoforms, regions of the protein that are prone to interact with protein partners, due to disorder-to-order transitions, here highlighted in red. The EDSCV motif found at the C-terminal of isoforms b and d matches the consensus sequence specifically recognized by class I PDZ domains. The other regions predicted in isoforms ‘a' and ‘d' may be recognized by unknown protein partners. Highlighted in red are the putative residues of binding motifs. (d) The exon 27 encoding for most of the M10 domain and the C-terminal tail where putative binding motifs are located. The KXD/E is highly conserved. Sequence of Homo sapiens was compared to the four species of mammals chosen (Ensembl database. ID: Homo Sapiens ATP2C1-011 ENST00000428331; Pan troglodytes ATP2C1-201 ENSPTRT00000028742; Canis lupus ATP2C1-201 ENSCAFT00000031759; Mus musculus ATP2C1-005 ENSMUST00000038118; Rattus norvegicus ATP2C1-201 ENSRNOT00000018175) and two non-mammalian (Ensembl database. ID: Gallus gallus ATP2C1-201 ENSGALT00000018903; Danio rerio ATP2C1-001 ENSDART00000084528)