Figure 7.
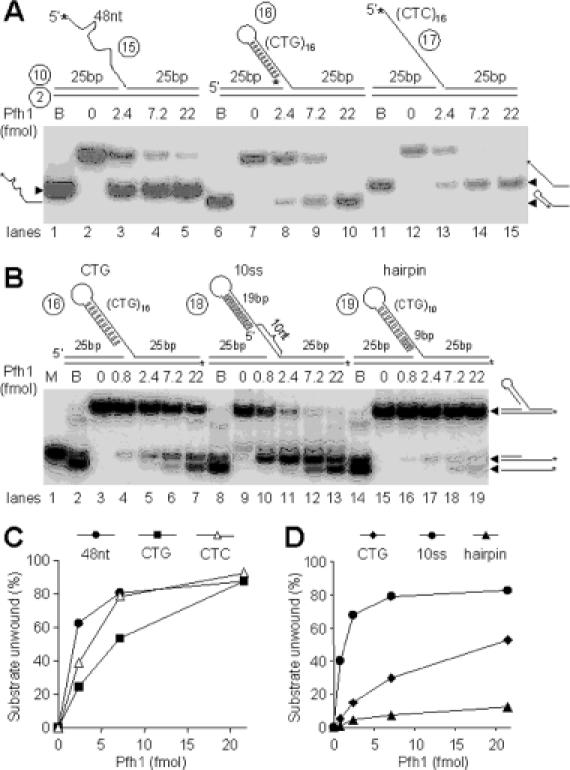
Pfh1 helicase can efficiently displace the hairpin-containing flap strand from DNA substrates. The schematic structures of substrates and oligonucleotides used are shown at the top of each figure. (A) The flap contains either 48 nt random sequence, (CTG)16, or (CTC)16 repeat at the 5′ tail. The asterisks indicate 32P-labeled ends. Increasing levels (0, 2.4, 7.2 and 22 fmol) of NusA-Pfh1-F in a 20 μl reaction mixture (see Materials and Methods) containing 100 mM NaCl were incubated with 15 fmol of each DNA substrate at 30°C for 10 min, and the products were analyzed on 10% polyacrylamide gel. B denotes boiled substrate controls. (B) The flap DNA substrates with a 10 nt or no gap between the hairpin and the junction are shown along with the (CTG)16 flap substrate at the top of the figure. The asterisks indicate 32P-labeled ends. Increasing amounts of Pfh1 (0, 0.8, 2.4, 7.2 and 22 fmol) in a 20 μl reaction mixture containing 100 mM NaCl were incubated at 30°C for 10 min with 15 fmol of each DNA substrate indicated at the top of figure, and the products were analyzed on 10% polyacrylamide gel. B and M denotes boiled substrate controls and size marker for intermediate substrate, respectively. (C) Quantification of the amount of substrate unwound in (A) as a function of Pfh1 added. (D) Quantification of the levels of the two products formed in (B) as a function of Pfh1 added.
