Figure 3.
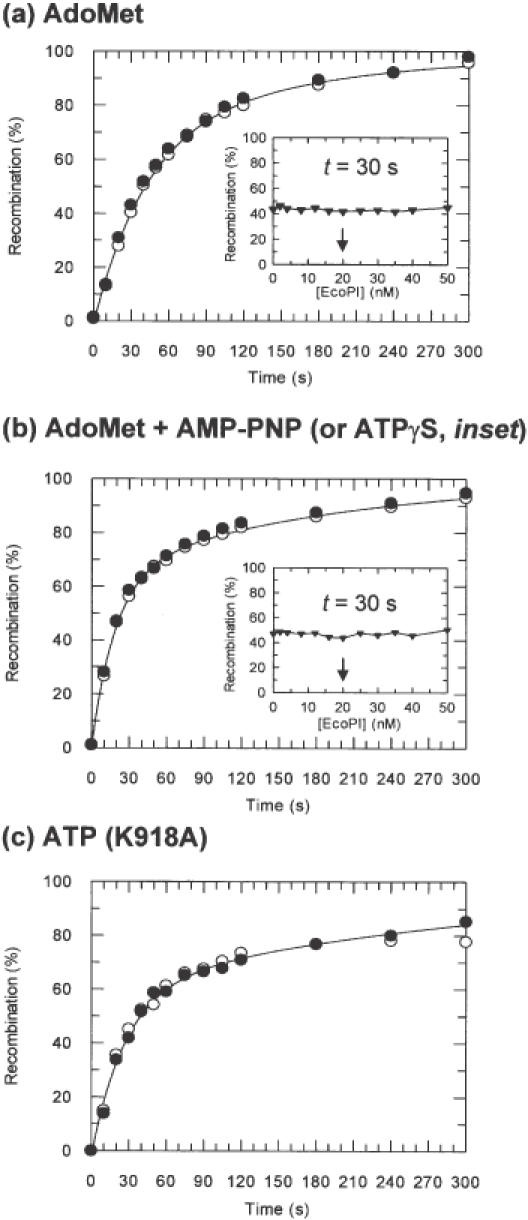
Tn21 resolvase recombination as a function of EcoPI binding. For kinetic reactions, 10 nM pMDS36a (20 nM Type III sites) was pre-incubated either with (black circles) or without (white circles) EcoPI (see Materials and Methods and text for further details). For titration experiments (black triangles), 10 nM pMDS36a was pre-incubated with EcoPI at the concentrations indicated (see Materials and Methods and text for further details). Curves represent least squares best fits of the data in the absence of EcoPI to a double exponential function (24). (A) Recombination time course in the presence of AdoMet and absence of NTP. Where added, EcoPI was at 20 nM. (a, inset) Extent of recombination after 30 s in the presence of AdoMet and absence of NTP following pre-incubation with EcoPI at the concentrations indicated. The arrow indicates where the Res2Mod2:site ratio is 1:1. (b) Recombination time course in the presence of AdoMet and AMP-PNP. Where added, EcoPI was at 20 nM. (b, inset) Extent of recombination after 30 s in the presence of AdoMet and ATPγS following pre-incubation with EcoPI at the concentrations indicated. The arrow indicates where the Res2Mod2:site ratio is 1:1. (c) Recombination time course in the presence of ATP alone. Where added, EcoPI K918A was at 40 nM.
