Figure 4.
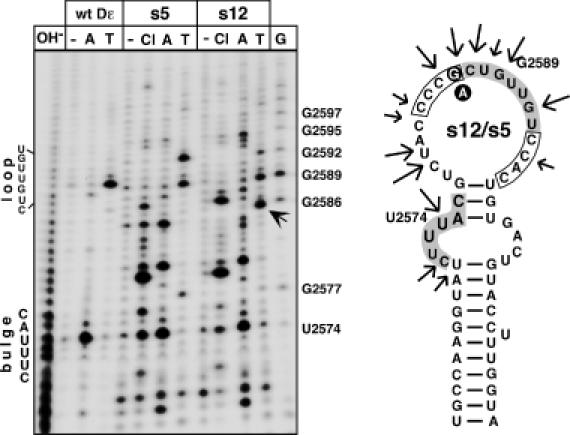
Secondary structure probing of round 9 aptamers s5 and s12. The 5′ abelled aptamers s12 (strongly priming) and s5 (weakly priming) were incubated with nuclease 3CL (Cl), RNase A (A), RNase T1 (T) or no enzyme (−). Reaction products were resolved by denaturing gel electrophoresis and visualized by autoradiography. The T1 product unique to s12 is highlighted by an arrow. Alkaline ladders (OH−) and G ladders (G) were obtained from wild-type Dε RNA. Positions of the bulge and loop in wild-type Dε are indicated on the left. On the secondary structure model the major nuclease sensitive sites are marked by arrows. The randomized regions are boxed, the bulge and loop residues as present in wild-type Dε are shown with grey background. The G versus an exchange is indicated by white letters on black background. The most stable M-Fold prediction has U2574 paired although it is highly accessible to RNase A; forcing it to be unpaired generates the structure shown. Note that both RNAs produce highly similar cleavage patterns. A corresponding analysis for aptamers s1 and s2 is shown as Supplementary Figure 4.
