Figure 5.
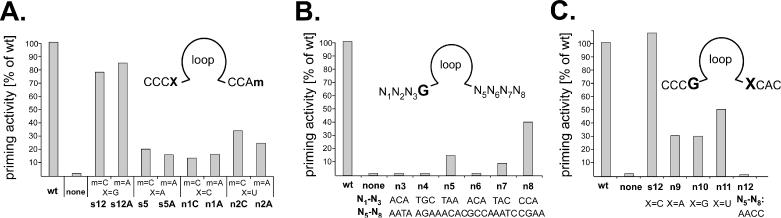
Priming activities of consensus-motif derived secondary generation aptamers. (A) Influence of the nucleotides at the N4 and N8 positions. Residue X (N4) in the consensus motif CCCX was changed from G to A, C and U, combined with either C or A at the m (N8) position in the downstream consensus motif CCAm. Nucleotides at the X and m positions, and the aptamer names are given below the graph; the X = G, m = C combination corresponds to s12. Bars show the relative priming signal intensities compared to wild-type Dε RNA set to 100%. (B) Context dependence of the priming-promoting activity of a G residue at N4. Aptamers with a G at N4 but various different sequences at N1 to N3 and N5 to N8 (shown below the graph) were subjected to in vitro priming assays. Note that only n8 with a sequence resembling the consensus motifs produced a significant signal. (C) Influence of the nucleotide at the N5 position. Within the priming-active CCCG–XCAC motif, the X residue (N5) was changed from C to A, G and U as indicated. Aptamer n12 is identical to s12A in (A), except that the sequence of the downstream CCAA motif is reversed. All priming assays shown in one graph were performed with one batch of P protein, and in parallel with positive (wild-type Dε RNA) and negative controls (no RNA).
