Figure 7.
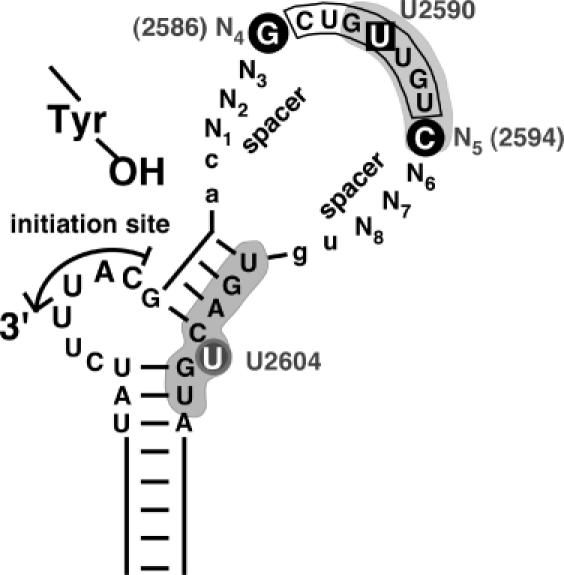
Revised view of relevant subelements in avian hepadnavirus ε signals. Based on this study and the expanded database of avian hepadnavirus sequences an open structure as in Hε, rather than Dε with its largely base paired upper stem, represents a prototypic ε signal. Functionally, the classical loop sequence (boxed) should be extended to encompass the adjacent N4 and N5 positions. N1 to N3 and N6 to N8, in contrast, serve mainly as spacers, with a lack of base pairing potential as a major requirement. Whether this also holds for the two preceding and the two following residues (lower case lettering) has not yet been experimentally addressed. Nucleotides with a specific role in priming are highlighted by black circles. Likely, they are part of a network of anchor residues, including the base pairs underlying the bulge and the unpaired U, that enable precise positioning of P over the initiation site. U2590 (boxed) in the loop sequence is one of the few residues whose mutation was previously shown to abrogate P-binding without affecting RNA secondary structure (18). Grey shading indicates a P protein footprint on the RNA (20). Notably, it includes the N5 position but spares N6 to N8 (and the two 3′ residues), in full accordance with the SELEX data and the natural sequence variability in this region.
