Abstract
The tumor suppressor p53 regulates cell-cycle progression and apoptosis in response to genotoxic stress, and inactivation of p53 is a common feature of cancer cells. The levels and activity of p53 are tightly regulated by posttranslational modifications, including phosphorylation, ubiquitination, and acetylation. Here, we demonstrate that the transcription factor Yin Yang 1 (YY1) interacts with p53 and inhibits its transcriptional activity. We show that YY1 disrupts the interaction between p53 and the coactivator p300 and that expression of YY1 blocks p300-dependent acetylation and stabilization of p53. Furthermore, expression of YY1 inhibits the accumulation of p53 and the induction of p53 target genes in response to genotoxic stress. YY1 also interacts with Mdm2 and the expression of YY1 promotes the assembly of the p53–Mdm2 complex. Consequently, YY1 enhances Mdm2-mediated ubiquitination of p53. Inactivation of endogenous YY1 enhances the accumulation of p53 as well as the expression of p53 target genes in response to DNA damage, and it sensitizes cells to DNA damage-induced apoptosis. Hence, our results demonstrate that YY1 regulates the transcriptional activity, acetylation, ubiquitination, and stability of p53 by inhibiting its interaction with the coactivator p300 and by enhancing its interaction with the negative regulator Mdm2. YY1 may, therefore, be an important negative regulator of the p53 tumor suppressor in response to genotoxic stress.
Keywords: p300, acetylation, ubiquitin, Mdm2, transcription
The tumor suppressor p53 regulates cell-cycle progression and apoptosis in response to genotoxic stress, and inactivation of p53 is a common feature of cancer cells (1–3). In stressed cells, p53 is targeted by a cascade of posttranslational modifications, including Ser and Thr phosphorylation and acetylation (4–6). These modifications stabilize the p53 protein and promote its translocation to the nucleus. p53 is a short-lived protein, and the amount and activity of p53 is maintained at low levels in normal cells. The ubiquitination and degradation of p53 are controlled mainly by Mdm2, an oncogenic ubiquitin ligase (7–10). Recently, it was suggested that Mdm2-mediated monoubiquitination of p53 inactivates the protein by promoting its nuclear export (11). Also, it has been suggested that the coactivator p300 could catalyze the polyubiquitination of p53 and, thereby, promote its degradation (12). However, it has also been demonstrated that p300-dependent acetylation of C-terminal lysine residues in p53 stabilizes the protein by preventing Mdm2-mediated ubiquitination of the same residues (13, 14). Similar mechanisms have also been reported for other proteins, including Smad7 and members of the SREBP family of transcription factors (15, 16). Acetylated p53 needs to be deacetylated before ubiquitination and degradation. Interestingly, it has been suggested that Mdm2 is involved in the recruitment of the proteins involved in the deacetylation of p53 (13). Thus, the balance between acetylation, deacetylation, ubiquitination, and deubiquitination will control the stability and function of p53 (4–6, 13, 14, 17–19). Proteins with intrinsic histone acetyltransferase activity act as transcriptional coactivators by acetylating histones, and thereby, they induce an open chromatin conformation, which allows the transcriptional machinery to have access to promoters (20). The best-characterized histone acetyltransferases are p300, CREB-binding protein (CBP), and P/CAF (20). These proteins interact with many transcription factors, including p53 (21, 22). Thus, it is believed that histone acetyltransferases are central integrators of various signaling pathways in the nucleus. Since the observation was made that p53 is a direct target for acetylation by the coactivators p300 and P/CAF (23), a number of transcription factors and nuclear proteins have been found to be modified in this manner (24–26). The functional consequences of p53 acetylation are diverse, and they include increased DNA-binding (23, 27), increased stability (13, 14), and enhanced interactions with coactivators (28). Here, we demonstrate that the transcription factor Yin Yang 1 (YY1) binds p53 and inhibits its transcriptional activity by interfering with the recruitment of p300. As a result, YY1 inhibits p300-mediated acetylation and stabilization of p53. YY1 also interacts with Mdm2 and promotes the formation of a p53–Mdm2 complex, thereby enhancing Mdm2-mediated ubiquitination of p53. Consequently, expression of YY1 inhibits the accumulation of active p53, as well as the expression of p53 target genes after DNA damage. Also, inactivation of endogenous YY1 sensitizes cells to DNA damage-induced apoptosis. Thus, our results identify YY1 as a regulator of the p53 tumor suppressor in response to genotoxic stress.
Methods
Antibodies, Plasmids, and Recombinant Adenoviruses. Antibodies directed against p53 (DO-1 and FL-393), Mdm2 (H-221), YY1 (H-414), Gal4-DBD (RK5C1), α-tubulin (TU-02), GADD45 (H-165), p21 (F-5), p300 (N-15), and hemagglutinin (HA; Y-11) were obtained from Santa Cruz Biotechnology. Anti-acetyl-histone H3 antibodies (catalog no. 06-599) were obtained from Upstate Biotechnology (Lake Placid, NY). Antibodies directed against phospho-p53 (Ser-15) (9284) were obtained from Cell Signaling Technology (Beverly, MA). Etoposide, doxycycline, and anti-Flag antibody (M5) were obtained from Sigma. The plasmids pcDNA3-YY1, pcDNA3-Flag-YY1, and pcDNA3-AS-YY1 have been described (29). The Gal4-YY1 constructs, as well as the Mdm2-luc, RGC-luc, and p21-luc promoter-reporter plasmids have been described (29–31). The plasmids pcDNA3-p53 and pcDNA3-Flag-p53, as well as the GST-p53 constructs, were generated by PCR amplification of human p53, followed by cloning into the appropriate expression vector. The YY1 adenovirus was generated by PCR amplification of human YY1 and cloning into the vector pAdTrack cytomegalovirus (32).
Cell Lines and Transfections. U2OS, H1299, HEK293, and HEK293T cells were obtained from the American Type Culture Collection. The primary human foreskin fibroblast cell line (AG01518) was obtained from Coriell Cell Repositories (Camden, NJ). p53-positive and p53-negative HCT116 cells were provided by B. Vogelstein (33). The Saos-2 TetOn p53-inducible cells (p53-TetOn) were provided by K. H. Vousden (Beatson Laboratories, Glasgow, Scotland) (34). Transient transfections in U2OS, H1299, HEK293, and HEK293T cells were performed by using the MBS transfection kit (Stratagene). p53-TetOn cells were transfected by using Effectene (Qiagen, Valencia, CA). HCT116 cells were transfected by using Lipifectamine 2000 (Invitrogen). For transcriptional assays, cells were transfected with the reporter gene in the absence or presence of expression vectors for the indicated proteins or empty expression vector (pcDNA3). The total amount of expression vector was constant in all transfections. After 36 h, luciferase activities were determined in duplicate samples, according to the manufacturer's instructions (Promega), by using cotransfected β-galactosidase as an internal control for transfection efficiency. The data represent the mean ± SD of three independent experiments performed in duplicates. The YY1 and control small-interfering RNA (siRNA) (GL2, directed against firefly luciferase) were obtained from Dharmacon (Lafayette, CO) and have been described (29).
Protein Analysis. Cells were lysed in buffer A (50 mM Hepes, pH 7.2/150 mM NaCl/1 mM EDTA/20 mM NaF/2 mM NaVO4 /10 mM β-glycerophosphate/1%, wt/vol, Triton X-100/10%, wt/vol, glycerol/1 mM PMSF/10 mM sodium butyrate/1% aprotinin/0.1% SDS/0.5% deoxycholate) and cleared by centrifugation. For coimmunoprecipitations, cell lysates were prepared in the absence of SDS and sodium deoxycholate. For ubiquitination assays, cell lysates were prepared under denaturing conditions and processed as described (15). Cell lysates and immunoprecipitates were resolved by SDS/PAGE and transferred to nitrocellulose membranes (Millipore). To ensure that equal amounts of protein were loaded in each well, the levels of tubulin in the samples were estimated by Western blotting using antitubulin antibodies. Proteins were translated in vitro by using the T7 TNT kit (Promega) with 35S-labeled methionine and cysteine, and they were incubated with GST-fusion proteins as described (15). The samples were washed extensively and resolved by SDS/PAGE. The gels were stained with Coommassie blue, dried, and analyzed by phosphorimage analysis.
Apoptosis Assays. Determination of cytoplasmic histone-associated DNA fragments was performed with the cell-death detection ELISA kit, according to the manufacturer's instructions (Roche). Caspase-dependent cleavage of cytokeratin 18 in methanol-fixed cells was detected with the M30 CytoDeath antibody by fluorescence microscopy according to the manufacturer's instructions (Roche). A total of 500 cells from four separate slides were counted in each experiment.
Chromatin Immunoprecipitation (ChIP) Assay. The ChIP assays were performed essentially as described (28). Immunoprecipitations were performed with 30 μg of DNA by using one of the following antibodies: 2 μg of anti-p53 (FL-393), 2 μg of anti-HA (control, Y-11), or 6 μg of anti-acetyl-histone H3. We used 1/10 of the immunoprecipitated material and 1/100 of the input DNA for analysis by PCR in the presence of 1 μCi (1 Ci = 37 GBq) α-[32P]dCTP. PCR products were resolved on 6% polyacrylamide gels and exposed to film. PCR amplification was performed with primers covering the 5′ p53-binding sequence within the human p21 promoter (forward, 5′ACCTTTCACCATTCCCCTAC; and reverse, 5′GCCCAAGGACAAAATAGCCA).
Results
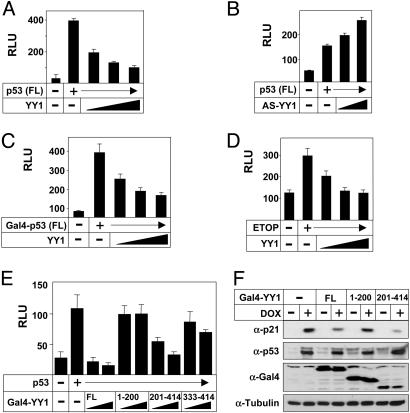
We picked up a number of proteins, including p53 and Smads, in a screen for transcription factors regulated in response to YY1 expression (29). To confirm the results of the initial screen, we performed p53-dependent transcriptional assays. p53-dependent activation of a Mdm2-luc promoter-reporter gene in U2OS cells was inhibited in response to YY1 expression (Fig. 1A). Similar results were also obtained in p53-negative H1299 and HCT116 cells (data not shown). Expression of an YY1 antisense construct in U2OS cells resulted in increased expression of Mdm2-luc in response to p53 (Fig. 1B), indicating that endogenous YY1 regulates p53. Expression of YY1 in U2OS cells also inhibited the transcriptional activity of Gal4-p53 on a promoter-reporter gene containing Gal4-binding sites (Fig. 1C), indicating that the effect of YY1 is independent of the intrinsic DNA-binding activity of p53. Expression of the p53-responsive RGC-luc promoter-reporter gene was enhanced in etoposide-treated U2OS cells, and the expression of YY1 blocked this effect (Fig. 1D), suggesting that YY1 interferes with the function of p53 in response to DNA damage. To identify the domain in YY1 that is required for inhibition of p53, fragments of YY1 were fused to the DNA-binding domain of Gal4 and the fusion proteins were used in p53-dependent transcriptional assays (Fig. 1E). Full-length YY1 inhibited p53-dependent expression of the Mdm2-luc promoter-reporter gene in U2OS cells. The same result was obtained with a construct containing a C-terminal portion of YY1 (amino acids 201–414), whereas both N-terminal (amino acids 1–200) and extreme C-terminal sequences (amino acids 333–414) were unable to inhibit p53. To test whether YY1 could inhibit p53-dependent expression of endogenous targets, we used p53-deficient Saos-2 cells engineered to express a p53 transgene under the control of a doxycycline-dependent promoter (p53-TetOn) (34). After transient transfection, Gal4-YY1 (full length) and Gal4-YY1 (amino acids 201–414) inhibited the expression of p21, whereas Gal4-YY1 (amino acids 1–200) was unable to do so (Fig. 1F). Together, our results suggest that YY1 inhibits the transcriptional activity of p53 and that the central region of YY1 (amino acids 201–333) is required for this effect.
Fig. 1.
YY1 inhibits the transcriptional activity of p53. (A) Activity of the Mdm2-luc reporter gene in U2OS cells transfected with p53 (25 ng) in the absence or presence of YY1 (pcDNA3-YY1, 20–100 ng) or empty vector (pcDNA3). FL, full length. (B) Activity of the Mdm2-luc reporter gene in U2OS cells transfected with p53 (25 ng) in the absence or presence of an antisense YY1 construct (pcDNA3-AS-YY1, 100–250 ng) or empty vector (pcDNA3). FL, full length. (C) Activity of the G5E1b-luc reporter gene in U2OS cells transfected with Gal4-p53 (25 ng) in the absence or presence of YY1 (pcDNA3-YY1, 20–100 ng) or empty vector (pcDNA3). FL, full length. (D) Activity of the RGC-luc reporter gene in U2OS cells transfected in the absence or presence of YY1 (pcDNA3-YY1, 100 ng) or empty vector (pcDNA3, 100 ng). Where indicated, cells were treated with etoposide (ETOP, 5 μM) 12 h before analysis. (E) Activity of Mdm2-luc reporter gene in U2OS cells transfected with p53 (25 ng) in the absence or presence of Gal4-YY1 (10 and 20 ng), either full-length (FL) or the indicated deletion mutants. (F) p53-TetOn cells were transfected with Gal4-YY1 (250 ng), either full-length (FL) or the indicated deletion mutants. Where indicated, the cells were treated with doxycycline (DOX, 200 μg/ml) for 12 h to induce p53 expression. The levels of p21, p53, Gal4-YY1 and α-tubulin were estimated by Western blot analysis.
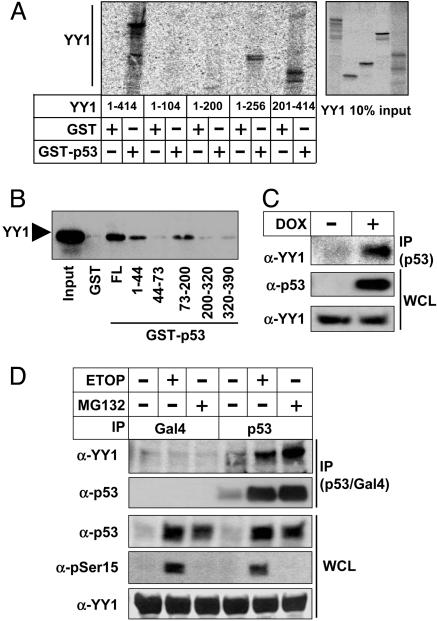
To determine whether p53 and YY1 interacted, fragments of YY1 were translated in vitro and used in GST-p53 pull-down assays. Full-length YY1 interacted with GST-p53 (Fig. 2A). Fragments containing the central region of YY1 also interacted with p53 (amino acids 1–256 and 201–414), whereas N-terminal fragments of YY1 were unable to interact. Similar results were also obtained in coimmunoprecipitation experiments of endogenous p53 and overexpressed Gal4-YY1 fragments (data not shown). These results correlate well with our transcriptional data (Fig. 1 E and F), demonstrating that the central region of YY1 is required for inhibition of p53. To identify the domains in p53 that interacted with YY1, full-length YY1 was expressed in 293T cells and used in GST-p53 pull-down assays by using different p53 domains fused to GST (Fig. 2B). Full-length YY1 interacted with full-length GST-p53. YY1 also interacted with the most N-terminal portion of the p53 transactivation domain (amino acids 1–44) and with a central portion of p53 (amino acids 73–200). Anti-p53 antibodies could only immunoprecipitate endogenous YY1 in p53-TetOn cells after doxycycline-dependent expression of p53 (Fig. 2C). Furthermore, endogenous YY1 coimmunoprecipitated with endogenous p53 in U2OS cells, and the interaction was enhanced after DNA damage-induced stabilization of p53 (Fig. 2D), whereas an unrelated control antibody was unable to immunoprecipitate YY1. However, stabilization of p53 with the proteasome inhibitor MG132 also enhanced the interaction between p53 and YY1, without having any effect on the phosphorylation of p53. Thus, the enhanced interaction between p53 and YY1 after DNA damage is probably independent of the phosphorylation of p53.
Fig. 2.
YY1 interacts with p53. (A) YY1, either full-length (1-414) or the indicated deletion mutants was translated in vitro and used in GST pull-down assays with GST alone or GST-p53. The amount of YY1 was analyzed by phosphorimage analysis. (B) Full-length YY1 was expressed in 293T cells and the cell lysates used in GST pull-downs with GST alone or the indicated GST-p53 proteins. FL, full length. The levels of YY1 were determined by Western blot analysis. (C) Cell lysates from p53-TetOn cells treated in the absence or presence of doxycycline (DOX) were immunoprecipitated (IP) with p53 antibodies (DO-1). Coimmunoprecipitated YY1 and the levels of p53 and YY1 in wholecell lysates (WCL) were determined by Western blot analysis. (D) Whole-cell lysates from U2OS cells treated in the absence or presence of etoposide (ETOP, 5 μM) or MG132 (25 μM) were immunoprecipitated (IP) with p53 antibodies (DO-1) or an unrelated antibody (anti-Gal4). Immunoprecipitated YY1 and p53 and the levels of p53 and YY1 and the phosphorylation of p53 on Ser-15 in whole-cell lysates (WCL) were determined by Western blot analysis.
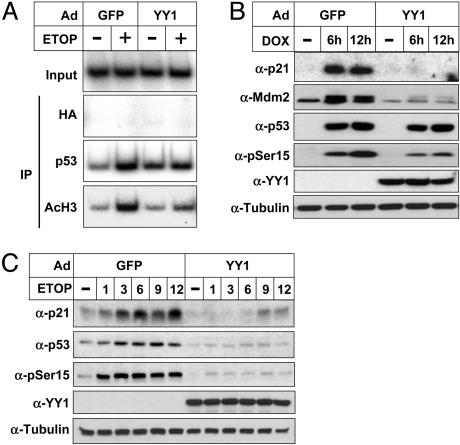
To assess the effect of YY1 on p53-dependent transcription in vivo, we used U2OS cells infected with adenoviruses expressing either GFP or YY1. The infected cells were treated in the absence or presence of etoposide to induce DNA damage, and they were used for ChIP assays (Fig. 3A). p53 was recruited to the p21 promoter in response to etoposide treatment in control cells, resulting in an enhanced acetylation of histone H3. In line with our previous results (Fig. 1), YY1 decreased the levels of both p53 and acetylated histone H3 associated with the p21 promoter. Doxycycline treatment of p53-TetOn cells resulted in the expression of the p53 transgene and induced a robust induction of the p21 and Mdm2 proteins in cells infected with the control virus (Fig. 3B). However, p53-dependent induction of both proteins was blocked in cells infected with the YY1 virus. To address whether YY1 could regulate the expression of p53 targets in response to genotoxic stress, U2OS cells were infected with adenoviruses expressing either GFP or YY1 and treated with etoposide to induce DNA damage. Etoposide treatment of control cells resulted in the phosphorylation (Ser-15) and stabilization of p53, as well as a robust induction of the p21 protein (Fig. 3C). Interestingly, no phosphorylation or stabilization of p53 in response to etoposide treatment was observed in cells expressing YY1. Consequently, we could not detect any induction of p21 in these cells, indicating that YY1 inhibits the activation of p53 in response to DNA damage. Similar results were obtained also in primary human fibroblasts in response to etoposide treatment and UV irradiation (see Fig. 6, which is published as supporting information on the PNAS web site), indicating that YY1 can inhibit the activation of p53 and the expression of downstream target genes in response to multiple stress signals.
Fig. 3.
YY1 inhibits the activation of p53 in response to genotoxic stress. (A) ChIP analysis of the p21 promoter after DNA damage. U2OS cells were infected with control (GFP) or YY1 adenovirus and either left untreated or treated with etoposide (ETOP, 5 μM) for 8 h. Cells were processed for ChIP analysis by using the indicated antibodies for immunoprecipitation (IP). (B) p53-TetOn cells were infected with control (GFP) or YY1 adenovirus and treated with doxycycline (DOX) for the indicated times to induce p53 expression. The expression of the p21, Mdm2, p53, and YY1 proteins and the phosphorylation of p53 on Ser-15 were determined by Western blot analysis. (C) U2OS cells were infected with control (GFP) or YY1 adenovirus and treated with etoposide (ETOP, 5 μM) for the indicated times. The expression of p21, p53, YY1, and α-tubulin and the phosphorylation of p53 on Ser-15 were determined by Western blot analysis.
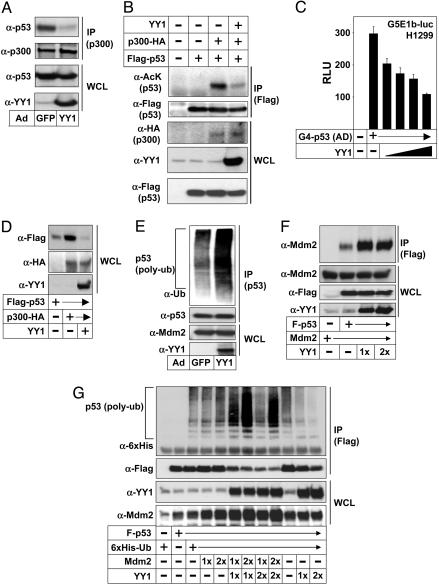
The acetyltransferases p300 and CBP regulate the function of p53 at several levels. Both proteins function as coactivators for p53-dependent transcription. Also, it has been demonstrated that p300-mediated acetylation of p53 enhances its DNA-binding activity, facilitates its interactions with other coactivators, and stabilizes the protein by preventing ubiquitination of the acetylated lysine residues. Interestingly, p53 and YY1 interact with the same domains in p300 and CBP (21, 22, 35). Thus, we hypothesized that YY1 could interfere with the interaction between p53 and p300 and, therefore, block p300-mediated acetylation of p53. To test this hypothesis, we analyzed the interaction between p53 and p300 in parental, p53-positive HCT116 cells after infection with either control or YY1 adenovirus. In support of our hypothesis, expression of YY1 blocked the interaction between endogenous p53 and p300 in HCT116 cells (Fig. 4A). Similar results were obtained when the three proteins were expressed in p53-deficient HCT116 and H1299 cells (data not shown). To further test our hypothesis, we analyzed p300-mediated acetylation of p53 in transfected p53-deficient HCT116 cells in the absence and presence of cotransfected YY1 (Fig. 4B). In line with our previous results, YY1 dramatically reduced the acetylation of p53 in response to p300 expression (Fig. 4B, compare lanes 3 and 4). p53 interacts with p300 and CBP through its N-terminal transactivation domain, and the transcriptional activity of this domain depends on these interactions. A fusion protein containing the DNA-binding domain of Gal4 and the transactivation domain of p53 was able to activate transcription from a minimal promoter containing Gal4-binding sites (Fig. 4C). In support of our hypothesis, YY1 inhibited the transcriptional activity of the p53 activation domain, suggesting that the interaction between this domain of p53 and p300 is an important target for YY1-mediated inhibition of the transcriptional activity of p53. It has been suggested that p300-mediated acetylation of p53 enhances its DNA-binding activity (23, 27, 36, 37). To test whether YY1 could affect these processes, p53-deficient H1299 cells were transfected with p53 in the absence or presence of p300 and YY1, and the recruitment of p53 to the p21 promoter was analyzed in ChIP assays (see Fig. 7, which is published as supporting information on the PNAS web site). Expression of p300 enhanced the association of p53 with the p21 promoter. Importantly, expression of YY1 reduced the p300-mediated recruitment of p53 to the promoter. Thus, our results demonstrate that YY1 inhibits p53-dependent transcription by interfering with the interaction between p53 and p300. By interfering with the interaction between p53 and p300, YY1 also inhibits p300-mediated acetylation p53.
Fig. 4.
YY1 interferes with the interaction between p53 and p300 and promotes Mdm2-mediated ubiquitination of p53. (A) p53-positive HCT116 cells were infected with control (GFP) or YY1 adenovirus. Endogenous p300 was immunoprecipitated (IP) from total cell lysates with anti-p300 antibodies. The amount of immunoprecipitated p53 and the levels of p53, p300, and YY1 in whole-cell lysates (WCL) were determined by Western blot analysis. (B) p53-deficient HCT116 cells were transfected with Flag-p53 (1 μg) in the absence or presence of p300-HA (2.5 μg) and YY1 (1 μg) or empty expression vector (pcDNA3). Flag-p53 was immunoprecipitated (IP) with Flag antibodies from whole-cell lysates. The acetylation of Flag-p53 was detected with anti-acetyl-lysine antibodies (α-AcK). The amount of immunoprecipitated Flag-p53 was determined by Western blot analysis. (C) The activity of the G5E1b-luc reporter gene was determined in H1299 cells transfected with Gal4-p53-AD (10 ng), a fusion protein containing the DNA-binding domain of Gal4 and the transactivation domain of p53 (amino acids 1–73), in the absence or presence of YY1 (20 and 40 ng) or empty expression plasmid (pcDNA3). (D) p53-negative HCT116 cells were transfected with Flag-p53 (0.1 μg) in the absence or presence of p300-HA (0.5 μg) and YY1 (0.25 μg) or empty expression vector (pcDNA3). The levels of Flag-p53, p300-HA, and YY1 in whole-cell lysates (WCL) were determined by Western blot analysis. (E) U2OS cells were infected with control (GFP) or YY1 adenovirus and treated with MG132 (25 μM) for 4 h before lysis. Whole-cell lysates were immunoprecipitated (IP) with anti-p53 antibodies, and the ubiquitination of p53 and the amount of p53 in the immunoprecipitates, as well as the levels of Mdm2 and YY1 in whole-cell lysates (WCL), were determined by Western blot analysis. (F) HEK293 cells were transfected with Mdm2 (0.5 μg) in the absence or presence of Flag-p53 (0.25 μg) and increasing amounts of YY1 (0.1 and 0.2 μg) or empty expression vector (pcDNA3). Whole-cell lysates were immunoprecipitated (IP) with anti-Flag antibodies, and coimmunoprecipitated Mdm2 was detected by Western blot analysis. The levels of Mdm2, Flag-p53, and YY1 in the whole-cell lysates (WCL) were determined by Western blot analysis. (G) HEK293 cells were transfected with Flag-p53 (0.25 μg) and Hisubiquitin (1 μg) in the absence or presence of Mdm2 (0.25 and 0.5) and YY1 (0.1 and 0.2 μg) or empty expression vector (pcDNA3). Whole-cell lysates were immunoprecipitated (IP) with anti-Flag antibodies and separated by SDS/PAGE. Ubiquitination of p53 and the amount of p53 in the immunoprecipitates, and the levels of Mdm2 and YY1 in whole-cell lysates (WCL) were determined by Western blot analysis.
The acetylated lysine residues in p53 are located in its C terminus, and it has been suggested that p300-mediated acetylation of these residues contributes to the stabilization of p53. To determine whether YY1 could overcome p300-mediated stabilization of p53, p53-deficient HCT116 cells were transfected with p53 in the absence and presence of p300 and YY1 (Fig. 4D). Expression of p300 enhanced the stability of the transfected p53 protein, and expression of YY1 inhibited this effect (Fig. 4D, compare lanes 2 and 3), demonstrating that YY1 can inhibit p300-mediated stabilization of p53. To test whether YY1 could affect the ubiquitination of endogenous p53, U2OS cells were infected with either control or YY1 adenovirus and the ubiquitination of p53 was analyzed after immunoprecipitation of the endogenous p53 protein. The ubiquitination of p53 was significantly enhanced in U2OS cells infected with the YY1 adenovirus (Fig. 4E), indicating that YY1 promotes the ubiquitination of p53 in vivo. The ubiquitination and degradation of p53 are largely controlled by the ubiquitin ligase Mdm2. YY1 interacted with Mdm2 after expression of the two proteins in HEK293 cells (see Fig. 8, which is published as supporting information on the PNAS web site). Interestingly, expression of YY1 enhanced the interaction between p53 and Mdm2 (Fig. 4F), indicating that YY1 may function as a scaffold protein to promote the assembly of the p53–Mdm2 complex. Importantly, Mdm2-dependent ubiquitination of p53 was enhanced in response to expression of YY1 in HEK293 cells (Fig. 4G), suggesting that YY1 promotes Mdm2-mediated ubiquitination and degradation of p53.
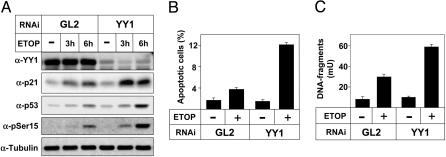
Our data indicate that YY1 negatively regulates the activation and transcriptional activity of p53 after DNA damage. Consequently, we wanted to determine whether YY1 could affect p53-mediated apoptosis. To address this issue, we used YY1-directed siRNA to inactivate endogenous YY1 in U2OS cells (Fig. 5A). The DNA damage-dependent phosphorylation and stabilization of p53 was enhanced in cells treated with YY1 siRNA compared with cells treated with control siRNA. Consequently, the expression of the p21 protein in response to etoposide treatment was enhanced in cells treated with YY1 siRNA. Similar results were obtained in MCF-7 cells (data not shown). When apoptosis was monitored in fixed U2OS cells with an antibody directed against cleaved cytokeratin 18 (M30), a 3-fold increase in apoptosis was observed after etoposide treatment of cells transfected with YY1 siRNA compared with cells treated with control siRNA (Fig. 5B). Similar results were also obtained with an antibody directed against cleaved caspase 3 (data not shown). When apoptosis in U2OS cells was monitored by determination of cytoplasmic histone-associated DNA fragments, a 2-fold increase in apoptosis was observed in response to etoposide treatment of cells transfected with YY1 siRNA compared with cells treated with control siRNA (Fig. 5C). Similar results were also obtained in p53-TetOn cells treated with YY1 siRNA (data not shown). Thus, our results demonstrate that endogenous YY1 interferes with the accumulation of active p53 in response to genotoxic stress, thereby inhibiting DNA damage-induced apoptosis.
Fig. 5.
Inactivation of YY1 promotes apoptosis in response to DNA damage. (A) siRNA-mediated inactivation of YY1. U2OS cells were transfected with control (GL2) or YY1 siRNA (20 nM) and treated with etoposide (5 μM) for the indicated times. The expression of p21, p53, YY1, and α-tubulin and the phosphorylation of p53 on Ser-15 were determined by Western blot analysis. (B) U2OS cells were transfected with control (GL2) or YY1 siRNA (20 nM) and treated with etoposide (5 μM) for 12 h. Apoptosis was determined after staining of fixed cells with an antibody directed against cleaved cytokeratin 18 (M30). (C) U2OS cells were transfected with siRNA and treated as described in B, and apoptosis was monitored by determination of cytoplasmic histone-associated DNA fragments by ELISA.
Discussion
YY1 is a multifunctional transcription factor, which is involved in transcriptional activation, repression, and initiation (29, 38–40). Targeted disruption of the YY1 gene leads to embryonic lethality in mice (41), indicating that YY1 plays critical roles during development. Here, we demonstrate that YY1 binds the p53 tumor suppressor and inhibits its transcriptional activity. Expression of YY1 inhibits the expression of p53 target genes after DNA damage. In addition, siRNA-mediated inactivation of endogenous YY1 sensitized cells to DNA damage-induced apoptosis. The acetyl-transferase p300 is an important regulator of the transcriptional activity of p53, and disruption of the p300 gene in HCT116 cells demonstrated that p300 also plays important roles for the function of p53 in vivo (42). YY1 and p53 interact with overlapping domains in p300, and we have demonstrated that YY1 interferes with the interaction between p53 and p300. Consequently, YY1 inhibited the transcriptional activity of the isolated activation domain of p53. However, p300 regulates the function of p53 through multiple pathways. It has been demonstrated that p53 is targeted by p300-mediated acetylation and that the acetylation of p53 is enhanced in response to various stress signals (4, 5). The acetylated lysine residues in p53 are located in its C terminus and coincide with residues that are important for the ubiquitination and degradation of the protein. It has been demonstrated that acetylation of these lysine residues stabilize p53 by interfering with Mdm2-mediated ubiquitination of the same residues (13, 14). In addition, it has been suggested that p300 has the ability to catalyze the polyubiquitination of p53 (12). Thus, p300 could be of major importance for the regulated degradation of p53. In support of this hypothesis, we found that expression of YY1 blocked the stabilization and accumulation of p53 in response to various stress signals. Furthermore, silencing of endogenous YY1 enhanced the stability of p53 after DNA damage. In addition, we could demonstrate that expression of YY1 inhibited p300-mediated acetylation and stabilization of p53. We also found that expression of YY1 enhanced the ubiquitination of endogenous p53. The ubiquitination of p53 is mainly controlled by Mdm2, an oncogenic ubiquitin ligase (7–10), and we could demonstrate that YY1 promoted Mdm2-mediated polyubiquitination of p53. In part, this effect could be explained by our observation that YY1 interacted with Mdm2 and enhanced the interaction between p53 and Mdm2. Alternatively, YY1 could promote the ubiquitination of p53 by preventing p300-mediated acetylation of C-terminal lysine residues, thus making them available for Mdm2-mediated ubiquitination. Thus, our results suggest that YY1 regulates the transcriptional activity, acetylation, ubiquitination, and stability of p53 by interfering with its interaction with p300 and promoting its interaction with Mdm2. However, the effects on the ubiquitination and stability of p53 are not absolutely required for YY1-mediated repression of the transcriptional activity of p53 because YY1 inhibited the transcriptional activity of the isolated activation domain of p53 when this portion of the protein was fused to the DNA-binding domain of Gal4. This fusion protein lacks the lysine residues targeted by Mdm2-mediated ubiquitination, and its transcriptional activity depends on the recruitment of p300 and other coactivators, suggesting that interference with coactivator recruitment is important for YY1-mediated inhibition of the transcriptional activity of p53. This hypothesis was supported by our observation that YY1 inhibited the p300-dependent recruitment of p53 to the p21 promoter in vivo. Similar results were reported recently for the immediate-early 2 protein (IE2) of human cytomegalovirus (37). It was demonstrated that IE2 inhibited the acetylation of p53 and, thereby, blocked the recruitment of p53 to its target promoters. Also, in p53-TetOn cells, in which the expression of p53 is controlled by a strong promoter, YY1 did not affect the steady-state levels of p53 significantly, yet the expression of downstream target genes was reduced (Figs. 1F and 3B). It is likely that YY1-mediated inhibition of p53 in vivo involves a combination of the effects reported in this study. First, YY1 inhibits the interaction between p53 and p300, thereby directly inhibiting the transcriptional activity of p53. By inhibiting the interaction between p53 and p300, YY1 also prevents p300-mediated acetylation of C-terminal lysine residues in p53, thereby making these residues available for Mdm2-mediated ubiquitination. Also, YY1 augments Mdm2-mediated ubiquitination of p53 by enhancing its interaction with Mdm2. During the preparation of this article, Yakovleva et al. (43) reported that recombinant YY1 and YY1 present in nuclear extracts could interact with a subset of p53 response elements in vitro. Thus, the authors suggested that the binding of YY1 to these DNA elements could regulate the expression of certain p53-dependent genes. Although it is possible that such a mechanism is active in vivo, it fails to explain YY1-mediated inhibition of the phosphorylation, acetylation, and accumulation of p53 in response to DNA damage. Also, the p53 response element in the human Mdm2 promoter contains no potential YY1-binding sites, although YY1 blocks p53-dependent expression of both endogenous Mdm2 and Mdm2 promoter-reporter genes. However, it is possible that binding of YY1 to certain p53-responsive promoters could affect the expression of the corresponding genes, suggesting that YY1 could regulate the function of p53 at multiple levels.
Inactivation of p53 is one of the hallmarks of cancer cells, and our results demonstrate that YY1 inhibits p53 function. An extensive analysis of the expression and function of YY1 in cancer cells has not, to our knowledge, been performed. However, the YY1 gene was identified recently in a screen for candidate genes involved in the development of acute myeloid leukemia (AML) (44). It was demonstrated that the murine myeloid leukemia virus was frequently integrated in the upstream YY1 promoter, thereby increasing the expression of the YY1 gene. In addition, the same study demonstrated that the expression of YY1 was enhanced in numerous cases of human AML. Further work is needed to investigate the role of YY1 in human tumorigenesis. However, YY1 and its interactions with p53, Mdm2, and p300 could be an attractive therapeutic target in cancer.
Note Added in Proof. Since the submission of the manuscript, Sui et al. (45) reported that YY1 interacted with p53 and Mdm2 and enhanced the Mdm2-mediated ubiquitination of p53.
Supplementary Material
Acknowledgments
We thank M. Atchison, M. Oren, B. Vogelstein, K. Vousden, and A. Moustakas for reagents; C.-H. Heldin, A. Sundqvist, and M. T. Bengoechea for comments and discussions. J.E. is a Research Fellow of the Royal Swedish Academy of Sciences through a grant from the Knut and Alice Wallenberg Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: YY1, Yin Yang 1; CBP, CREB-binding protein; siRNA, small-interfering RNA; HA, hemagglutinin; ChIP, chromatin immunoprecipitation.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 2.Vousden, K. H. (2000) Cell 103, 691-694. [DOI] [PubMed] [Google Scholar]
- 3.Vousden, K. H. & Lu, X. (2002) Nat. Rev. Cancer 2, 594-604. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi, K., Herrera, J. E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C. W. & Appella, E. (1998) Genes Dev. 12, 2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, C. L. & Gu, W. (2003) Curr. Opin. Cell Biol. 15, 164-171. [DOI] [PubMed] [Google Scholar]
- 6.Appella, E. & Anderson, C. W. (2001) Eur. J. Biochem. 268, 2764-2772. [DOI] [PubMed] [Google Scholar]
- 7.Michael, D. & Oren, M. (2003) Semin. Cancer Biol. 13, 49-58. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. (1997) Nature 387, 299-303. [DOI] [PubMed] [Google Scholar]
- 9.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296-299. [DOI] [PubMed] [Google Scholar]
- 10.Yap, D. B., Hsieh, J. K. & Lu, X. (2000) J. Biol. Chem. 275, 37296-37302. [DOI] [PubMed] [Google Scholar]
- 11.Li, M., Brooks, C. L., Wu-Baer, F., Chen, D., Baer, R. & Gu, W. (2003) Science 302, 1972-1975. [DOI] [PubMed] [Google Scholar]
- 12.Grossman, S. R., Deato, M. E., Brignone, C., Chan, H. M., Kung, A. L., Tagami, H., Nakatani, Y. & Livingston, D. M. (2003) Science 300, 342-344. [DOI] [PubMed] [Google Scholar]
- 13.Ito, A., Kawaguchi, Y., Lai, C. H., Kovacs, J. J., Higashimoto, Y., Appella, E. & Yao, T. P. (2002) EMBO J. 21, 6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, M., Luo, J., Brooks, C. L. & Gu, W. (2002) J. Biol. Chem. 277, 50607-50611. [DOI] [PubMed] [Google Scholar]
- 15.Grönroos, E., Hellman, U., Heldin, C. H. & Ericsson, J. (2002) Mol. Cell 10, 483-493. [DOI] [PubMed] [Google Scholar]
- 16.Giandomenico, V., Simonsson, M., Grönroos, E. & Ericsson, J. (2003) Mol. Cell. Biol. 23, 2587-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J. & Gu, W. (2002) Nature 416, 648-653. [DOI] [PubMed] [Google Scholar]
- 18.Lim, S. K., Shin, J. M., Kim, Y. S. & Baek, K. H. (2004) Int. J. Oncol. 24, 357-364. [PubMed] [Google Scholar]
- 19.Luo, J., Su, F., Chen, D., Shiloh, A. & Gu, W. (2000) Nature 408, 377-381. [DOI] [PubMed] [Google Scholar]
- 20.Roth, S. Y., Denu, J. M. & Allis, C. D. (2001) Annu. Rev. Biochem. 70, 81-120. [DOI] [PubMed] [Google Scholar]
- 21.Grossman, S. R., Perez, M., Kung, A. L., Joseph, M., Mansur, C., Xiao, Z. X., Kumar, S., Howley, P. M. & Livingston, D. M. (1998) Mol. Cell 2, 405-415. [DOI] [PubMed] [Google Scholar]
- 22.Van Orden, K., Giebler, H. A., Lemasson, I., Gonzales, M. & Nyborg, J. K. (1999) J. Biol. Chem. 274, 26321-26328. [DOI] [PubMed] [Google Scholar]
- 23.Gu, W. & Roeder, R. G. (1997) Cell 90, 595-606. [DOI] [PubMed] [Google Scholar]
- 24.Bannister, A. J. & Miska, E. A. (2000) Cell. Mol. Life Sci. 57, 1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger, S. L. (1999) Curr. Opin. Cell Biol. 11, 336-341. [DOI] [PubMed] [Google Scholar]
- 26.Kouzarides, T. (2000) EMBO J. 19, 1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, J., Li, M., Tang, Y., Laszkowska, M., Roeder, R. G. & Gu, W. (2004) Proc. Natl. Acad. Sci. USA 101, 2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barlev, N. A., Liu, L., Chehab, N. H., Mansfield, K., Harris, K. G., Halazonetis, T. D. & Berger, S. L. (2001) Mol. Cell 8, 1243-1254. [DOI] [PubMed] [Google Scholar]
- 29.Kurisaki, K., Kurisaki, A., Valcourt, U., Terentiev, A. A., Pardali, K., Ten Dijke, P., Heldin, C. H., Ericsson, J. & Moustakas, A. (2003) Mol. Cell. Biol. 23, 4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushmeyer, S., Park, K. & Atchison, M. L. (1995) J. Biol. Chem. 270, 30213-30220. [DOI] [PubMed] [Google Scholar]
- 31.Zauberman, A., Flusberg, D., Haupt, Y., Barak, Y. & Oren, M. (1995) Nucleic Acids Res. 23, 2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J.P., Sedivy, J.M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497-1501. [DOI] [PubMed] [Google Scholar]
- 34.Ryan, K. M., Ernst, M. K., Rice, N. R. & Vousden, K. H. (2000) Nature 404, 892-897. [DOI] [PubMed] [Google Scholar]
- 35.Austen, M., Luscher, B. & Luscher-Firzlaff, J. M. (1997) J. Biol. Chem. 272, 1709-1717. [DOI] [PubMed] [Google Scholar]
- 36.Dornan, D., Shimizu, H., Burch, L., Smith, A.J. & Hupp, T. R. (2003) Mol. Cell. Biol. 23, 8846-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu, C. H., Chang, M. D., Tai, K. Y., Yang, Y. T., Wang, P. S., Chen, C. J., Wang, Y. H., Lee, S. C., Wu, C. W. & Juan L. J. (2004) EMBO J. 23, 2269-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galvin, K. M. & Shi, Y. (1997) Mol. Cell. Biol. 17, 3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, M. J. & Seto, E. (1999) Gene 236, 197-208. [DOI] [PubMed] [Google Scholar]
- 40.Atchison, L., Ghias, A., Wilkinson, F., Bonini, N. & Atchison, M. L. (2003) EMBO J. 22, 1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donohoe, M. E., Zhang, X., McGinnis, L., Biggers, J., Li, E. & Shi, Y. (1999) Mol. Cell. Biol. 19, 7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer, N. G., Chin, S. F., Ozdag, H., Daigo, Y., Hu, D. E., Cariati, M., Brindle, K., Aparicio, S. & Caldas, C. (2004) Proc. Natl. Acad. Sci. USA 101, 7386-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yakovleva, T., Kolesnikova, L., Vukojevic, V., Gileva, I., Tan-No, K., Austen, M., Luscher, B., Ekstrom, T. J., Terenius, L. & Bakalkin, G. (2004) Biochem. Biophys. Res. Commun. 318, 615-624. [DOI] [PubMed] [Google Scholar]
- 44.Erkeland, S. J., Valkhof, M., Heijmans-Antonissen, C., Delwel, R., Valk, P. J., Hermans, M. H. & Touw, I. P. (2003) Blood 101, 1111-1117. [DOI] [PubMed] [Google Scholar]
- 45.Sui, G., Affar el, B., Shi, Y., Brignone, C., Wall, N. R., Yin, P., Donohoe, M., Luke, M. P. Calvor D., Grossman, S. R. & Shi, Y. (2004) Cell 117, 859-872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.