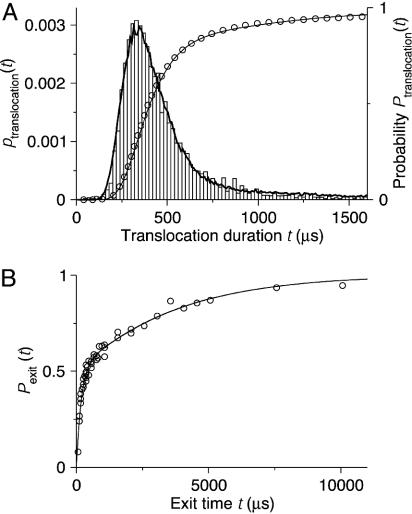
Fig. 6.
DNA translocation experiment of Bates et al. (5) modeled by the trap-diffusion model. (A) Probability distributions of poly(dA)60 translocation time t across α-hemolysin channels at 120 mV. The experimental results (bars and open circles for probability density and cumulative distribution, respectively; data taken from figure 2 of ref. 5) are compared with Brownian dynamics simulations of the trap-diffusion model [Eqs. 1 and 2 (lines)]. In the simulations, t was estimated as the first-passage time to drift downstream by a distance L (where L is an arbitrary scaling length corresponding here to the extension of the DNA) with parameters D = 1.35 × 10-4 L2/μs, v = 2.5 × 10-3 L/μs, kon = 1/(1,750 μs), and koff = 1/(600 μs). (B) Cumulative probability distributions of exit times. Experimental results, shown as open circles, are taken from figure 5 of ref. 5. In the simulations (line), as in the experiments, the “polymer” was drawn into the pore for 200 μs at 120 mV with the parameters of A, before the electric field was turned off and the cumulative exit-time distribution was collected (with DNA exit at both ends of the pore). In the field-free period, we used D = 3.6 × 10-4 L2/μs, v = 0, kon = 1/(450 μs), and koff = 1/(1,900 μs).